A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates
- PMID: 25219855
- PMCID: PMC4209185
- DOI: 10.1038/nature13723
A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates
Abstract
A defining feature governing head patterning of jawed vertebrates is a highly conserved gene regulatory network that integrates hindbrain segmentation with segmentally restricted domains of Hox gene expression. Although non-vertebrate chordates display nested domains of axial Hox expression, they lack hindbrain segmentation. The sea lamprey, a jawless fish, can provide unique insights into vertebrate origins owing to its phylogenetic position at the base of the vertebrate tree. It has been suggested that lamprey may represent an intermediate state where nested Hox expression has not been coupled to the process of hindbrain segmentation. However, little is known about the regulatory network underlying Hox expression in lamprey or its relationship to hindbrain segmentation. Here, using a novel tool that allows cross-species comparisons of regulatory elements between jawed and jawless vertebrates, we report deep conservation of both upstream regulators and segmental activity of enhancer elements across these distant species. Regulatory regions from diverse gnathostomes drive segmental reporter expression in the lamprey hindbrain and require the same transcriptional inputs (for example, Kreisler (also known as Mafba), Krox20 (also known as Egr2a)) in both lamprey and zebrafish. We find that lamprey hox genes display dynamic segmentally restricted domains of expression; we also isolated a conserved exonic hox2 enhancer from lamprey that drives segmental expression in rhombomeres 2 and 4. Our results show that coupling of Hox gene expression to segmentation of the hindbrain is an ancient trait with origin at the base of vertebrates that probably led to the formation of rhombomeric compartments with an underlying Hox code.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
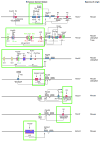


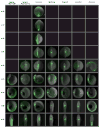


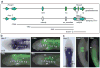

References
-
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. - PubMed
-
- Shimeld SM, Donoghue PC. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish) Development. 2012;139:2091–2099. - PubMed
-
- Murakami Y, et al. Segmental development of reticulospinal and branchiomotor neurons in lamprey: insights into the evolution of the vertebrate hindbrain. Development. 2004;131:983–995. - PubMed
-
- Takio Y, et al. Evolutionary biology: lamprey Hox genes and the evolution of jaws. Nature. 2004;429:1–262. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

