Class I HDAC inhibition stimulates cardiac protein SUMOylation through a post-translational mechanism
- PMID: 25220405
- PMCID: PMC4252807
- DOI: 10.1016/j.cellsig.2014.09.005
Class I HDAC inhibition stimulates cardiac protein SUMOylation through a post-translational mechanism
Abstract
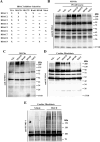
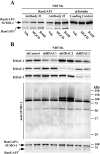
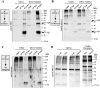
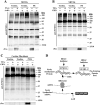
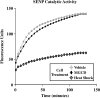
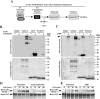
Lysine residues are subject to a multitude of reversible post-translational modifications, including acetylation and SUMOylation. In the heart, enhancement of lysine acetylation or SUMOylation using histone deacetylase (HDAC) inhibitors or SUMO-1 gene transfer, respectively, has been shown to be cardioprotective. Here, we addressed whether there is crosstalk between lysine acetylation and SUMOylation in the heart. Treatment of cardiomyocytes and cardiac fibroblasts with pharmacological inhibitors of HDAC catalytic activity robustly increased conjugation of SUMO-1, but not SUMO-2/3, to several high molecular weight proteins in both cell types. The use of a battery of selective HDAC inhibitors and short hairpin RNAs demonstrated that HDAC2, which is a class I HDAC, is the primary HDAC isoform that controls cardiac protein SUMOylation. HDAC inhibitors stimulated protein SUMOylation in the absence of de novo gene transcription or protein synthesis, revealing a post-translational mechanism of HDAC inhibitor action. HDAC inhibition did not suppress the activity of de-SUMOylating enzymes, suggesting that increased protein SUMOylation in HDAC inhibitor-treated cells is due to stimulation of SUMO-1 conjugation rather than blockade of SUMO-1 cleavage. Consistent with this, multiple components of the SUMO conjugation machinery were capable of being acetylated in vitro. These findings reveal a novel role for reversible lysine acetylation in the control of SUMOylation in the heart, and suggest that cardioprotective actions of HDAC inhibitors are in part due to stimulation of protein SUMO-1-ylation in myocytes and fibroblasts.
Keywords: Acetylation; HDAC; SUMO.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

References
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

