Efflux pump proteins in antifungal resistance
- PMID: 25221515
- PMCID: PMC4148622
- DOI: 10.3389/fphar.2014.00202
Efflux pump proteins in antifungal resistance
Abstract
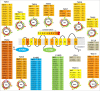
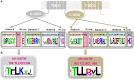
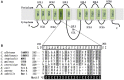
It is now well-known that the enhanced expression of ATP binding cassette (ABC) and major facilitator superfamily (MFS) proteins contribute to the development of tolerance to antifungals in yeasts. For example, the azole resistant clinical isolates of the opportunistic human fungal pathogen Candida albicans show an overexpression of Cdr1p and/or CaMdr1p belonging to ABC and MFS superfamilies, respectively. Hence, azole resistant isolates display reduced accumulation of therapeutic drug due to its rapid extrusion and that facilitates its survival. Considering the importance of major antifungal transporters, the focus of recent research has been to understand the structure and function of these proteins to design inhibitors/modulators to block the pump protein activity so that the drug already in use could again sensitize resistant yeast cells. The review focuses on the structure and function of ABC and MFS transporters of Candida to highlight the recent advancement in the field.
Keywords: ABC transporters; Candida; MFS transporters; azoles; efflux pumps; multidrug resistance.
Figures



References
-
- Ananthaswamy N., Rutledge R., Sauna Z. E., Dine E., Nelson E., Xia D., et al. (2010). The signaling interface of the yeast multidrug transporter Pdr5 adopts a cis conformation and there are functional overlap and equivalence of the deviant and canonical Q-loop residues. Biochemistry 49, 4440–4449 10.1021/bi100394j - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

