Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine
- PMID: 25222836
- PMCID: PMC4281956
- DOI: 10.1186/s13059-014-0453-8
Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine
Abstract
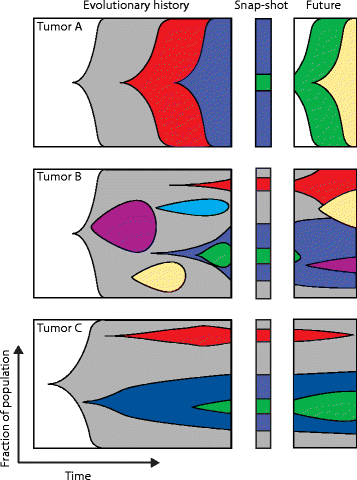
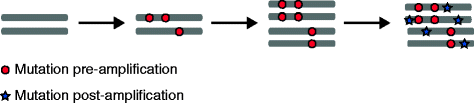
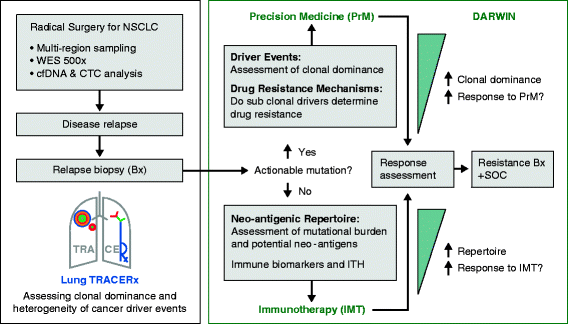
The presence of multiple subclones within tumors mandates understanding of longitudinal and spatial subclonal dynamics. Resolving the spatial and temporal heterogeneity of subclones with cancer driver events may offer insight into therapy response, tumor evolutionary histories and clinical trial design.
Figures

References
-
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

