Unraveling the sequence-dependent polymorphic behavior of d(CpG) steps in B-DNA
- PMID: 25223784
- PMCID: PMC4191396
- DOI: 10.1093/nar/gku809
Unraveling the sequence-dependent polymorphic behavior of d(CpG) steps in B-DNA
Abstract
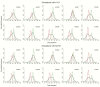
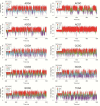
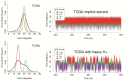
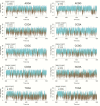
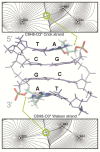
We have made a detailed study of one of the most surprising sources of polymorphism in B-DNA: the high twist/low twist (HT/LT) conformational change in the d(CpG) base pair step. Using extensive computations, complemented with database analysis, we were able to characterize the twist polymorphism in the d(CpG) step in all the possible tetranucleotide environment. We found that twist polymorphism is coupled with BI/BII transitions, and, quite surprisingly, with slide polymorphism in the neighboring step. Unexpectedly, the penetration of cations into the minor groove of the d(CpG) step seems to be the key element in promoting twist transitions. The tetranucleotide environment also plays an important role in the sequence-dependent d(CpG) polymorphism. In this connection, we have detected a previously unexplored intramolecular C-H···O hydrogen bond interaction that stabilizes the low twist state when 3'-purines flank the d(CpG) step. This work explains a coupled mechanism involving several apparently uncorrelated conformational transitions that has only been partially inferred by earlier experimental or theoretical studies. Our results provide a complete description of twist polymorphism in d(CpG) steps and a detailed picture of the molecular choreography associated with this conformational change.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Franklin R.E., Gosling R.G. Molecular configuration in sodium thymonucleate. 1953. Nature. 2003;421:400–401. discussion 396. - PubMed
-
- Radhakrishnan I., Patel D.J. DNA triplexes: solution structures, hydration sites, energetics, interactions, and function. Biochemistry. 1994;33:11405–11416. - PubMed
-
- Bernués J., Azorín F. Triple-stranded DNA. In: Eckstein F., Lilley D.J., editors. Nucleic Acids and Molecular Biology SE - 1. Springer Berlin Heidelberg: Nucleic Acids and Molecular Biology; 1995. pp. 1–21.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

