GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots
- PMID: 25224536
- PMCID: PMC4172956
- DOI: 10.1186/s12870-014-0245-z
GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots
Abstract
Background: Promoters play important roles in gene expression and function. There are three basic types of promoters: constitutive, specific, and inducible. Constitutive promoters are widely used in genetic engineering, but these promoters have limitations. Inducible promoters are activated by specific inducers. Tissue-specific promoters are a type of specific promoters that drive gene expression in specific tissues or organs. Here, we cloned and characterized the GmPRP2 promoter from soybean. The expression pattern indicated that this promoter is root-preferential in transgenic Arabidopsis and the hairy roots of soybean. It can be used to improve the root resistance or tolerance to pathogens, pests, malnutrition and other abiotic stresses which cause extensive annual losses in soybean production.
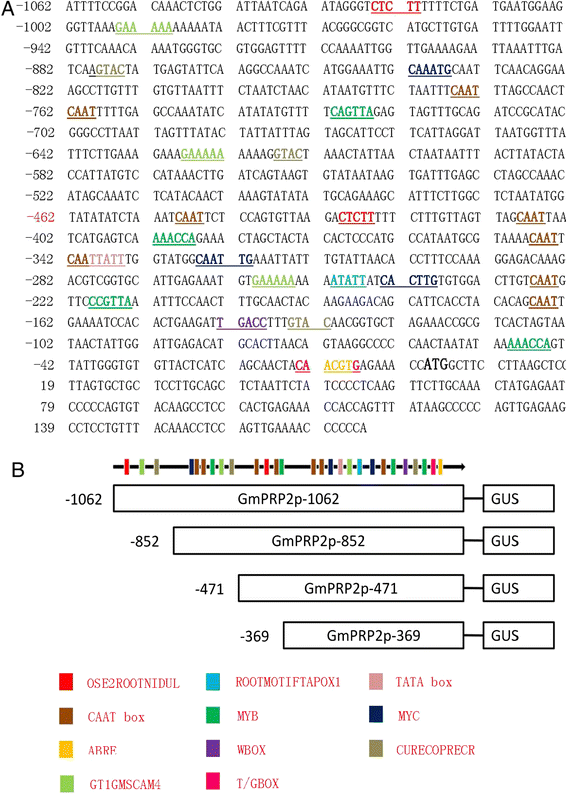
Results: The GmPRP2 promoter (GmPRP2p-1062) was isolated from soybean cv. Williams 82. Sequence analysis revealed that this promoter contains many cis-acting elements, including root-specific motifs. The GmPRP2p-1062 and its 5'-deletion fragments were fused with the GUS reporter gene and introduced into Arabidopsis and the hairy roots of soybean to further determine promoter activity. Histochemical analysis in transgenic Arabidopsis showed that GUS activity was mainly detected in roots and hypocotyls in all deletion fragments except GmPRP2p-471 (a 5'-deletion fragment of GmPRP2p-1062 with 471 bp length). GUS activity was higher in transgenic Arabidopsis and hairy roots with GmPRP2p-1062 and GmPRP2p-852 (a 5'-deletion fragment of GmPRP2p-1062 with 852 bp length) constructs than the other two constructs. GUS activity was enhanced by NaCl, PEG, IAA and JM treatments and decreased by SA, ABA and GA treatments in transgenic Arabidopsis.
Conclusions: GmPRP2p-1062 is a root-preferential promoter, and its core fragment for root-preferential expression might lie between -369 and +1. GmPRP2p-852 may be useful in the genetic engineering of novel soybean cultivars in the future.
Figures







Similar articles
-
Analysis of expression characteristics of soybean leaf and root tissue-specific promoters in Arabidopsis and soybean.Transgenic Res. 2021 Dec;30(6):799-810. doi: 10.1007/s11248-021-00266-7. Epub 2021 Jun 11. Transgenic Res. 2021. PMID: 34115286
-
Tinkering Cis Motifs Jigsaw Puzzle Led to Root-Specific Drought-Inducible Novel Synthetic Promoters.Int J Mol Sci. 2020 Feb 18;21(4):1357. doi: 10.3390/ijms21041357. Int J Mol Sci. 2020. PMID: 32085397 Free PMC article.
-
Identification of a 193 bp promoter region of TaNRX1-D gene from common wheat that contributes to osmotic or ABA stress inducibility in transgenic Arabidopsis.Genes Genomics. 2021 Sep;43(9):1035-1048. doi: 10.1007/s13258-021-01115-x. Epub 2021 Jun 18. Genes Genomics. 2021. PMID: 34143419
-
Overexpression of an aquaporin protein from Aspergillus glaucus confers salt tolerance in transgenic soybean.Transgenic Res. 2021 Dec;30(6):727-737. doi: 10.1007/s11248-021-00280-9. Epub 2021 Aug 30. Transgenic Res. 2021. PMID: 34460070 Review.
-
Mechanisms Underlying Soybean Response to Phosphorus Deficiency through Integration of Omics Analysis.Int J Mol Sci. 2022 Apr 21;23(9):4592. doi: 10.3390/ijms23094592. Int J Mol Sci. 2022. PMID: 35562981 Free PMC article. Review.
Cited by
-
The Roles of GmERF135 in Improving Salt Tolerance and Decreasing ABA Sensitivity in Soybean.Front Plant Sci. 2019 Jul 23;10:940. doi: 10.3389/fpls.2019.00940. eCollection 2019. Front Plant Sci. 2019. PMID: 31396249 Free PMC article.
-
Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.).Int J Mol Sci. 2018 Jul 10;19(7):2009. doi: 10.3390/ijms19072009. Int J Mol Sci. 2018. PMID: 29996483 Free PMC article.
-
Genetic engineering of parthenocarpic tomato plants using transient SlIAA9 knockdown by novel tissue-specific promoters.Sci Rep. 2019 Dec 11;9(1):18871. doi: 10.1038/s41598-019-55400-7. Sci Rep. 2019. PMID: 31827210 Free PMC article.
-
A Brief Review of Plant Cell Transfection, Gene Transcript Expression, and Genotypic Integration for Enhancing Compound Production.Methods Mol Biol. 2023;2575:153-179. doi: 10.1007/978-1-0716-2716-7_8. Methods Mol Biol. 2023. PMID: 36301475 Review.
-
Cell Wall Proteins Play Critical Roles in Plant Adaptation to Phosphorus Deficiency.Int J Mol Sci. 2019 Oct 23;20(21):5259. doi: 10.3390/ijms20215259. Int J Mol Sci. 2019. PMID: 31652783 Free PMC article. Review.
References
-
- Bhullar S, Chakravarthy S, Advani S, Datta S, Pental D, Burma PK. Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping. Plant Physiol. 2003;132:988–998. doi: 10.1104/pp.103.020602. - DOI - PMC - PubMed
-
- Charrier B, Scollan C, Ross S, Zubko E, Meyer P. Co-silencing of homologous transgenes in tobacco. Mol Breed. 2000;6:407–419. doi: 10.1023/A:1009672714835. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

