Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis
- PMID: 25225609
- PMCID: PMC4164332
- DOI: 10.1186/2049-2618-2-29
Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis
Abstract
Background: Changes in respiratory tract microbiota have been associated with diseases such as tuberculosis, a global public health problem that affects millions of people each year. This pilot study was carried out using sputum, oropharynx, and nasal respiratory tract samples collected from patients with pulmonary tuberculosis and healthy control individuals, in order to compare sample types and their usefulness in assessing changes in bacterial and fungal communities.
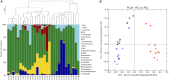
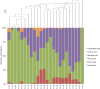
Findings: Most V1-V2 16S rRNA gene sequences belonged to the phyla Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria, with differences in relative abundances and in specific taxa associated with each sample type. Most fungal ITS1 sequences were classified as Ascomycota and Basidiomycota, but abundances differed for the different samples. Bacterial and fungal community structures in oropharynx and sputum samples were similar to one another, as indicated by several beta diversity analyses, and both differed from nasal samples. The only difference between patient and control microbiota was found in oropharynx samples for both bacteria and fungi. Bacterial diversity was greater in sputum samples, while fungal diversity was greater in nasal samples.
Conclusions: Respiratory tract microbial communities were similar in terms of the major phyla identified, yet they varied in terms of relative abundances and diversity indexes. Oropharynx communities varied with respect to health status and resembled those in sputum samples, which are collected from tuberculosis patients only due to the difficulty in obtaining sputum from healthy individuals, suggesting that oropharynx samples can be used to analyze community structure alterations associated with tuberculosis.
Keywords: 16S rRNA; ITS1; Microbial diversity; Microbiota; Mycobacterium tuberculosis; Pulmonary tuberculosis; Respiratory tract.
Figures
References
-
- Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One. 2012;7:e36313. - PMC - PubMed
-
- Zhou Y, Lin P, Li Q, Han L, Zheng H, Wei Y, Cui Z, Ni Y, Guo X. Analysis of the microbiota of sputum samples from patients with lower respiratory tract infections. Acta Biochim Biophys Sin (Shanghai) 2010;42:754–761. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

