A monovalent cation acts as structural and catalytic cofactor in translational GTPases
- PMID: 25225612
- PMCID: PMC4283411
- DOI: 10.15252/embj.201488517
A monovalent cation acts as structural and catalytic cofactor in translational GTPases
Abstract
Translational GTPases are universally conserved GTP hydrolyzing enzymes, critical for fidelity and speed of ribosomal protein biosynthesis. Despite their central roles, the mechanisms of GTP-dependent conformational switching and GTP hydrolysis that govern the function of trGTPases remain poorly understood. Here, we provide biochemical and high-resolution structural evidence that eIF5B and aEF1A/EF-Tu bound to GTP or GTPγS coordinate a monovalent cation (M(+)) in their active site. Our data reveal that M(+) ions form constitutive components of the catalytic machinery in trGTPases acting as structural cofactor to stabilize the GTP-bound "on" state. Additionally, the M(+) ion provides a positive charge into the active site analogous to the arginine-finger in the Ras-RasGAP system indicating a similar role as catalytic element that stabilizes the transition state of the hydrolysis reaction. In sequence and structure, the coordination shell for the M(+) ion is, with exception of eIF2γ, highly conserved among trGTPases from bacteria to human. We therefore propose a universal mechanism of M(+)-dependent conformational switching and GTP hydrolysis among trGTPases with important consequences for the interpretation of available biochemical and structural data.
Keywords: GTPase; catalytic mechanism; crystal structure; monovalent cation; translation.
© 2014 The Authors.
Figures
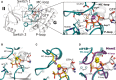
Overview of G domain and domain II of GTP-bound eIF5B with a Na+ ion (purple sphere) in the active site. P-loop and switch regions are shown in cyan; GTP is shown as balls and sticks. The Na+ ion is bound by a pentameric coordination sphere (inset; indicated by purple lines) formed by GTP, AspMC and GlyMC. Mg2+ and water molecules are shown as spheres in light brown and gray, respectively; the catalytic histidine (His480) and highly conserved residues that interact with GTP or the cations are indicated; hydrogen bonds are shown as dashed lines.
The active site of eIF5B·GTPγS with a K+ ion (yellow sphere) bound in a heptameric coordination sphere.
Superposition of eIF5B·GTPγS structures with either Na+ (purple sphere) or K+ (yellow sphere) bound in the active site. Due to the shorter coordination distances to the Na+ ion, AspMC and GlyMC (purple sticks) are drawn closer to the GTP molecule than in the K+-structure (yellow sticks).
Superposition of eIF5B·GTPγS·K+ and MnmE (purple; PDB: 2GJ8) bound to GDP, AlF4 and K+.


Excerpt of a multiple sequence alignment of different trGTPases (orthologs of RF3, eIF5B, eIF2γ, SelB, eRF3, EF-Tu, LepA, EF-G) showing P-loop and switch 1. The upper and lower numbering corresponds to C. thermophilum eIF5B (Cthe) and Escherichia coli EF-Tu (Ecoli), respectively. Highly conserved residues are highlighted in dark blue, conserved residues in light blue. AspMC and GlyMC are highlighted in red; residues in eIF2γ that replace AspMC and GlyMC are highlighted in yellow.
Superposition of eIF5B·GTPγS·K+ (colored as in Fig 1) with ribosome-bound EF-G·GDPCP (brown; PDB: 4JUW). Ribosome-bound EF-G provides all structural elements to bind the M+ ion; however, its coordination is prevented by the CH2 group of GDPCP in lieu of the β-γ-bridging oxygen (arrow). A water molecule (red sphere) is bound next to the M+-binding site instead.
Similarly, EF-Tu·GDPNP (purple; PDB: 2C78) provides all structural elements to bind the M+ ion; however, its coordination is prevented by the NH group of GDPNP (arrow). A water molecule (red sphere) is bound next to the M+ binding site instead.
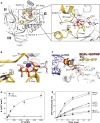
Overview of domains I (G) to III of GTP-bound aEF1A with a Na+ ion (purple sphere) in the active site. P-loop and switch regions are shown in yellow. The inset shows a detailed view on the active site with the coordination sphere of the Na+ ion (indicated by purple lines) formed by GTP, AspMC and GlyMC in the MC-loop.
Superposition of GTP-bound aEF1A with either a Na+ (purple sphere) or NH4+ ion (blue sphere) in the active site. Both ions are coordinated by the identical sphere, however, with different coordination distances.
Superposition of EF-Tu·GDPNP (switch 1 in red; PDB: 1EXM) with EF-Tu·GDPNP in the ternary complex (TC) with aa-tRNA (blue; PDBs: 1TTT, 1OB2) and aEF1A·GTP·M+ (M+ in blue; switch 1 in yellow). The M+ ion stabilizes a conformation of helix A″ that is required for stable TC formation.
Dependency of the intrinsic GTPase activity of E. coli EF-Tu wild-type (•) or the D21A mutant (○) on the concentration of K+ ions, determined in the presence of 0–1 M KCl under single turnover conditions (20 μM GTP-bound EF-Tu) at 30°C and subsequent analysis by HPLC.
Intrinsic GTPase activity of E. coli EF-Tu determined in the presence of 200 mM of the indicated salts under single turnover conditions. The order in which the combinations are given on the right corresponds to the relative rates of GTP hydrolysis.

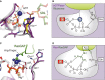
The nucleotide-dependent conformational switch in the G domain of a/eIF5B. In eIF5B·GDP, switch 1 and 2 are oriented away from the nucleotide binding pocket. Upon exchange of GDP for GTP, the switch regions undergo a large conformational rearrangement that results in direct contacts with the γ-phosphate. Here, the M+ ion (blue sphere) provides a direct contribution to the stabilization of switch 1 (inset). This contribution is missing in aIF5B·GDPNP (PDB: 1G7T), allowing it to crystallize in the GDP-like “off” state conformation.
Schematic presentation of the M+-dependent conformational switch mechanism in trGTPases.
Similar articles
-
Review: Translational GTPases.Biopolymers. 2016 Aug;105(8):463-75. doi: 10.1002/bip.22832. Biopolymers. 2016. PMID: 26971860 Free PMC article. Review.
-
Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome.J Mol Biol. 2003 Sep 19;332(3):689-99. doi: 10.1016/s0022-2836(03)00947-1. J Mol Biol. 2003. PMID: 12963376
-
The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation.Structure. 1993 Sep 15;1(1):35-50. doi: 10.1016/0969-2126(93)90007-4. Structure. 1993. PMID: 8069622
-
The conformation of a catalytic loop is central to GTPase activity on the ribosome.Biochemistry. 2015 Jan 20;54(2):546-56. doi: 10.1021/bi501373g. Epub 2014 Dec 30. Biochemistry. 2015. PMID: 25515218
-
Peculiarities in Activation of Hydrolytic Activity of Elongation Factors.Biochemistry (Mosc). 2020 Nov;85(11):1422-1433. doi: 10.1134/S0006297920110103. Biochemistry (Mosc). 2020. PMID: 33280582 Review.
Cited by
-
Common Mechanism of Activated Catalysis in P-loop Fold Nucleoside Triphosphatases-United in Diversity.Biomolecules. 2022 Sep 22;12(10):1346. doi: 10.3390/biom12101346. Biomolecules. 2022. PMID: 36291556 Free PMC article.
-
A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate.Sci Rep. 2015 Aug 12;5:12970. doi: 10.1038/srep12970. Sci Rep. 2015. PMID: 26264741 Free PMC article.
-
Role of aIF5B in archaeal translation initiation.Nucleic Acids Res. 2022 Jun 24;50(11):6532-6548. doi: 10.1093/nar/gkac490. Nucleic Acids Res. 2022. PMID: 35694843 Free PMC article.
-
Altered tRNA dynamics during translocation on slippery mRNA as determinant of spontaneous ribosome frameshifting.Nat Commun. 2022 Jul 22;13(1):4231. doi: 10.1038/s41467-022-31852-w. Nat Commun. 2022. PMID: 35869111 Free PMC article.
-
Saccharomyces cerevisiae Ski7 Is a GTP-Binding Protein Adopting the Characteristic Conformation of Active Translational GTPases.Structure. 2015 Jul 7;23(7):1336-43. doi: 10.1016/j.str.2015.04.018. Epub 2015 Jun 4. Structure. 2015. PMID: 26051716 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(2):213–221. Pt. - PMC - PubMed
-
- Ash MR, Maher MJ, Mitchell Guss J, Jormakka M. The cation-dependent G-proteins: in a class of their own. FEBS Lett. 2012;586:2218–2224. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

