Genome-wide identification and expression profiling analysis of the Aux/IAA gene family in Medicago truncatula during the early phase of Sinorhizobium meliloti infection
- PMID: 25226164
- PMCID: PMC4166667
- DOI: 10.1371/journal.pone.0107495
Genome-wide identification and expression profiling analysis of the Aux/IAA gene family in Medicago truncatula during the early phase of Sinorhizobium meliloti infection
Abstract
Background: Auxin/indoleacetic acid (Aux/IAA) genes, coding a family of short-lived nuclear proteins, play key roles in wide variety of plant developmental processes, including root system regulation and responses to environmental stimulus. However, how they function in auxin signaling pathway and symbiosis with rhizobial in Medicago truncatula are largely unknown. The present study aims at gaining deeper insight on distinctive expression and function features of Aux/IAA family genes in Medicago truncatula during nodule formation.
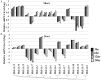
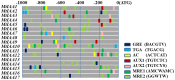
Principal findings: Using the latest updated draft of the full Medicago truncatula genome, a comprehensive identification and analysis of IAA genes were performed. The data indicated that MtIAA family genes are distributed in all the M. truncatula chromosomes except chromosome 6. Most of MtIAA genes are responsive to exogenous auxin and express in tissues-specific manner. To understand the biological functions of MtIAA genes involved in nodule formation, quantitative real-time polymerase chain reaction (qRT-PCR) was used to test the expression profiling of MtIAA genes during the early phase of Sinorhizobium meliloti (S. meliloti) infection. The expression patterns of most MtIAA genes were down-regulated in roots and up-regulated in shoots by S. meliloti infection. The differences in expression responses between roots and shoots caused by S. meliloti infection were alleviated by 1-NOA application.
Conclusion: The genome-wide identification, evolution and expression pattern analysis of MtIAA genes were performed in this study. The data helps us to understand the roles of MtIAA-mediated auxin signaling in nodule formation during the early phase of S. meliloti infection.
Conflict of interest statement
Figures






References
-
- Ljung K (2013) Auxin metabolism and homeostasis during plant development. Development 140: 943–950. - PubMed
-
- Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, et al. (2011) Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66: 564–578. - PubMed
-
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

