Morphological and population genomic evidence that human faces have evolved to signal individual identity
- PMID: 25226282
- PMCID: PMC4257785
- DOI: 10.1038/ncomms5800
Morphological and population genomic evidence that human faces have evolved to signal individual identity
Abstract
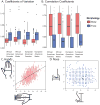
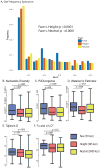
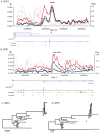
Facial recognition plays a key role in human interactions, and there has been great interest in understanding the evolution of human abilities for individual recognition and tracking social relationships. Individual recognition requires sufficient cognitive abilities and phenotypic diversity within a population for discrimination to be possible. Despite the importance of facial recognition in humans, the evolution of facial identity has received little attention. Here we demonstrate that faces evolved to signal individual identity under negative frequency-dependent selection. Faces show elevated phenotypic variation and lower between-trait correlations compared with other traits. Regions surrounding face-associated single nucleotide polymorphisms show elevated diversity consistent with frequency-dependent selection. Genetic variation maintained by identity signalling tends to be shared across populations and, for some loci, predates the origin of Homo sapiens. Studies of human social evolution tend to emphasize cognitive adaptations, but we show that social evolution has shaped patterns of human phenotypic and genetic diversity as well.
Conflict of interest statement
Figures

References
-
- Dunbar RIM, Shultz S. Evolution in the social brain. Science. 2007;317:1344–1347. - PubMed
-
- Byrne RW, Whiten A. Machiavellian Intelligence: Social Expertise and the Evolution of Intellect in Monkeys, Apes, and Humans. Oxford: Univeristy Press; 1988.

