Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib
- PMID: 25228534
- PMCID: PMC4233168
- DOI: 10.1158/1078-0432.CCR-14-1511
Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib
Abstract
Purpose: The first-generation ALK tyrosine kinase inhibitor (TKI) crizotinib is a standard therapy for patients with ALK-rearranged non-small cell lung cancer (NSCLC). Several next-generation ALK-TKIs have entered the clinic and have shown promising activity in crizotinib-resistant patients. As patients still relapse even on these next-generation ALK-TKIs, we examined mechanisms of resistance to the next-generation ALK-TKI alectinib and potential strategies to overcome this resistance.
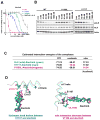
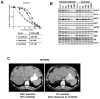
Experimental design: We established a cell line model of alectinib resistance, and analyzed a resistant tumor specimen from a patient who had relapsed on alectinib. We developed Ba/F3 models harboring alectinib-resistant ALK mutations and evaluated the potency of other next-generation ALK-TKIs in these models. We tested the antitumor activity of the next-generation ALK-TKI ceritinib in the patient with acquired resistance to alectinib. To elucidate structure-activity relationships of ALK mutations, we performed computational thermodynamic simulation with MP-CAFEE.
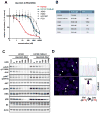
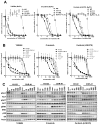
Results: We identified a novel V1180L gatekeeper mutation from the cell line model and a second novel I1171T mutation from the patient who developed resistance to alectinib. Both ALK mutations conferred resistance to alectinib as well as to crizotinib, but were sensitive to ceritinib and other next-generation ALK-TKIs. Treatment of the patient with ceritinib led to a marked response. Thermodynamics simulation suggests that both mutations lead to distinct structural alterations that decrease the binding affinity with alectinib.
Conclusions: We have identified two novel ALK mutations arising after alectinib exposure that are sensitive to other next-generation ALK-TKIs. The ability of ceritinib to overcome alectinib-resistance mutations suggests a potential role for sequential therapy with multiple next-generation ALK-TKIs.
©2014 American Association for Cancer Research.
Conflict of interest statement
Engelman JA.: Consultant for Genentech, Chugai, Novartis. Sponsored research from Novartis
Shaw AT.: Consultant for: Pfizer, Novartis, Chugai, Ariad, Ignyta, Daiichi-sankyo, and Genentech
Figures

Comment in
-
Perfect ALKemy: optimizing the use of ALK-directed therapies in lung cancer.Clin Cancer Res. 2014 Nov 15;20(22):5576-8. doi: 10.1158/1078-0432.CCR-14-2306. Epub 2014 Sep 16. Clin Cancer Res. 2014. PMID: 25228532 Free PMC article.
References
-
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. - PubMed
-
- Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

