The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription
- PMID: 25228644
- PMCID: PMC4173158
- DOI: 10.1101/gad.250225.114
The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription
Abstract
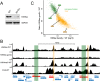
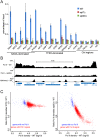
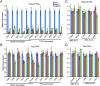
The SAGA (Spt-Ada-Gcn5 acetyltransferase) coactivator complex contains distinct chromatin-modifying activities and is recruited by DNA-bound activators to regulate the expression of a subset of genes. Surprisingly, recent studies revealed little overlap between genome-wide SAGA-binding profiles and changes in gene expression upon depletion of subunits of the complex. As indicators of SAGA recruitment on chromatin, we monitored in yeast and human cells the genome-wide distribution of histone H3K9 acetylation and H2B ubiquitination, which are respectively deposited or removed by SAGA. Changes in these modifications after inactivation of the corresponding enzyme revealed that SAGA acetylates the promoters and deubiquitinates the transcribed region of all expressed genes. In agreement with this broad distribution, we show that SAGA plays a critical role for RNA polymerase II recruitment at all expressed genes. In addition, through quantification of newly synthesized RNA, we demonstrated that SAGA inactivation induced a strong decrease of mRNA synthesis at all tested genes. Analysis of the SAGA deubiquitination activity further revealed that SAGA acts on the whole transcribed genome in a very fast manner, indicating a highly dynamic association of the complex with chromatin. Thus, our study uncovers a new function for SAGA as a bone fide cofactor for all RNA polymerase II transcription.
Keywords: RNA polymerase II; SAGA; acetyltransferase; chromatin; deubiquitinase; transcription.
© 2014 Bonnet et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
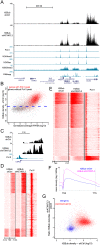
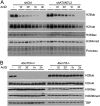
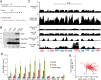
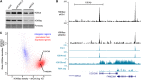
References
-
- Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
