Interlocked DNA nanostructures controlled by a reversible logic circuit
- PMID: 25229207
- PMCID: PMC4199106
- DOI: 10.1038/ncomms5940
Interlocked DNA nanostructures controlled by a reversible logic circuit
Abstract
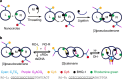
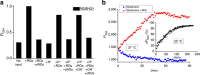
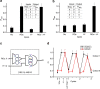
DNA nanostructures constitute attractive devices for logic computing and nanomechanics. An emerging interest is to integrate these two fields and devise intelligent DNA nanorobots. Here we report a reversible logic circuit built on the programmable assembly of a double-stranded (ds) DNA [3]pseudocatenane that serves as a rigid scaffold to position two separate branched-out head-motifs, a bimolecular i-motif and a G-quadruplex. The G-quadruplex only forms when preceded by the assembly of the i-motif. The formation of the latter, in turn, requires acidic pH and unhindered mobility of the head-motif containing dsDNA nanorings with respect to the central ring to which they are interlocked, triggered by release oligodeoxynucleotides. We employ these features to convert the structural changes into Boolean operations with fluorescence labelling. The nanostructure behaves as a reversible logic circuit consisting of tandem YES and AND gates. Such reversible logic circuits integrated into functional nanodevices may guide future intelligent DNA nanorobots to manipulate cascade reactions in biological systems.
Figures



Similar articles
-
Single-Molecule Manipulation of the Duplex Formation and Dissociation at the G-Quadruplex/i-Motif Site in the DNA Nanostructure.ACS Nano. 2015 Oct 27;9(10):9922-9. doi: 10.1021/acsnano.5b03413. Epub 2015 Sep 24. ACS Nano. 2015. PMID: 26371377
-
Logic gating by macrocycle displacement using a double-stranded DNA [3]rotaxane shuttle.Angew Chem Int Ed Engl. 2014 Sep 22;53(39):10372-6. doi: 10.1002/anie.201405447. Epub 2014 Jul 30. Angew Chem Int Ed Engl. 2014. PMID: 25078433
-
A DNA origami nanorobot controlled by nucleic acid hybridization.Small. 2014 Jul 23;10(14):2918-26. doi: 10.1002/smll.201400245. Epub 2014 Mar 20. Small. 2014. PMID: 24648163
-
DNA switches: from principles to applications.Angew Chem Int Ed Engl. 2015 Jan 19;54(4):1098-129. doi: 10.1002/anie.201404652. Epub 2014 Dec 17. Angew Chem Int Ed Engl. 2015. PMID: 25521588 Review.
-
From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures.Chem Rev. 2014 Mar 12;114(5):2881-941. doi: 10.1021/cr400354z. Epub 2014 Feb 27. Chem Rev. 2014. PMID: 24576227 Review. No abstract available.
Cited by
-
The effect of the neutral cytidine protonated analogue pseudoisocytidine on the stability of i-motif structures.Sci Rep. 2017 Jun 5;7(1):2772. doi: 10.1038/s41598-017-02723-y. Sci Rep. 2017. PMID: 28584239 Free PMC article.
-
I-Motif/miniduplex hybrid structures bind benzothiazole dyes with unprecedented efficiencies: a generic light-up system for label-free DNA nanoassemblies and bioimaging.Nucleic Acids Res. 2020 Feb 28;48(4):1681-1690. doi: 10.1093/nar/gkaa020. Nucleic Acids Res. 2020. PMID: 31950160 Free PMC article.
-
A DNA nanoswitch-controlled reversible nanosensor.Nucleic Acids Res. 2017 Jan 25;45(2):541-546. doi: 10.1093/nar/gkw1146. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899631 Free PMC article.
-
Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation.Nat Chem. 2017 Nov;9(11):1056-1067. doi: 10.1038/nchem.2852. Epub 2017 Sep 25. Nat Chem. 2017. PMID: 29064489 Free PMC article. Review.
-
Enzyme-free synthesis of cyclic single-stranded DNA constructs containing a single triazole, amide or phosphoramidate backbone linkage and their use as templates for rolling circle amplification and nanoflower formation.Chem Sci. 2018 Aug 24;9(42):8110-8120. doi: 10.1039/c8sc02952k. eCollection 2018 Nov 14. Chem Sci. 2018. PMID: 30542561 Free PMC article.
References
-
- Mao C., Sun W., Shen Z. & Seeman N. C. A nanomechanical device based on the B-Z transition of DNA. Nature 397, 144–146 (1999). - PubMed
-
- Yurke B., Turberfield A. J., Mills A. P. Jr., Simmel F. C. & Neumann J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000). - PubMed
-
- Liu D. & Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. Engl. 42, 5734–5736 (2003). - PubMed
-
- Chen Y. & Mao C. Putting a brake on an autonomous DNA nanomotor. J. Am. Chem. Soc. 126, 8626–8627 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

