IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies
- PMID: 25229428
- PMCID: PMC4226722
- DOI: 10.18632/oncotarget.2405
IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies
Abstract
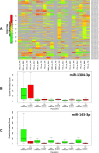
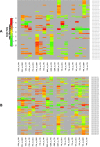
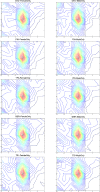
For many years it was believed that each mature microRNA (miRNA) existed as a single entity with fixed endpoints and a 'static' and unchangeable primary sequence. However, recent evidence suggests that mature miRNAs are more 'dynamic' and that each miRNA precursor arm gives rise to multiple isoforms, the isomiRs. Here we report on our identification of numerous and abundant isomiRs in the lymphoblastoid cell lines (LCLs) of 452 men and women from five different population groups. Unexpectedly, we find that these isomiRs exhibit an expression profile that is population-dependent and gender-dependent. This is important as it indicates that the LCLs of each gender/population combination have their own unique collection of mature miRNA transcripts. Moreover, each identified isomiR has its own characteristic abundance that remains consistent across biological replicates indicating that these are not degradation products. The primary sequences of identified isomiRs differ from the known miRBase miRNA either at their 5´-endpoint (leads to a different 'seed' sequence and suggests a different targetome), their 3´-endpoint, or both simultaneously. Our analysis of Argonaute PAR-CLIP data from LCLs supports the association of many of these newly identified isomiRs with the Argonaute silencing complex and thus their functional roles through participation in the RNA interference pathway.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Rigoutsos I, Tsirigos A. MicroRNA Target Prediction. In: Slack F, editor. MicroRNAs in Development and Cancer: Imperial College Press. 2010. pp. 237–259.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

