Signs of neutralization in a redundant gene involved in homologous recombination in Wolbachia endosymbionts
- PMID: 25230723
- PMCID: PMC4224334
- DOI: 10.1093/gbe/evu207
Signs of neutralization in a redundant gene involved in homologous recombination in Wolbachia endosymbionts
Abstract
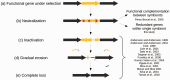
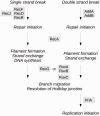
Genomic reduction in bacterial endosymbionts occurs through large genomic deletions and long-term accumulation of mutations. The latter process involves successive steps including gene neutralization, pseudogenization, and gradual erosion until complete loss. Although many examples of pseudogenes at various levels of degradation have been reported, neutralization cases are scarce because of the transient nature of the process. Gene neutralization may occur due to relaxation of selection in nonessential genes, for example, those involved in redundant functions. Here, we report an example of gene neutralization in the homologous recombination (HR) pathway of Wolbachia, a bacterial endosymbiont of arthropods and nematodes. The HR pathway is often depleted in endosymbiont genomes, but it is apparently intact in some Wolbachia strains. Analysis of 12 major HR genes showed that they have been globally under strong purifying selection during the evolution of Wolbachia strains hosted by arthropods, supporting the evolutionary importance of the HR pathway for these Wolbachia genomes. However, we detected signs of recent neutralization of the ruvA gene in a subset of Wolbachia strains, which might be related to an ancestral, clade-specific amino acid change that impaired DNA-binding activity. Strikingly, RuvA is part of the RuvAB complex involved in branch migration, whose function overlaps with the RecG helicase. Although ruvA is experiencing neutralization, recG is under strong purifying selection. Thus, our high phylogenetic resolution suggests that we identified a rare example of targeted neutralization of a gene involved in a redundant function in an endosymbiont genome.
Keywords: endosymbiosis; gene loss; genomic reduction; molecular evolution; nonorthologous gene displacement; selection relaxation.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Akman L, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andersson JO, Andersson SG. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. - PubMed
-
- Andersson JO, Andersson SG. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol Biol Evol. 2001;18:829–839. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

