Structural plasticity of the coiled-coil domain of rotavirus NSP4
- PMID: 25231315
- PMCID: PMC4248954
- DOI: 10.1128/JVI.02227-14
Structural plasticity of the coiled-coil domain of rotavirus NSP4
Abstract
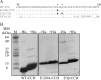
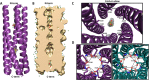
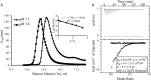
Rotavirus (RV) nonstructural protein 4 (NSP4) is a virulence factor that disrupts cellular Ca(2+) homeostasis and plays multiple roles regulating RV replication and the pathophysiology of RV-induced diarrhea. Although its native oligomeric state is unclear, crystallographic studies of the coiled-coil domain (CCD) of NSP4 from two different strains suggest that it functions as a tetramer or a pentamer. While the CCD of simian strain SA11 NSP4 forms a tetramer that binds Ca(2+) at its core, the CCD of human strain ST3 forms a pentamer lacking the bound Ca(2+) despite the residues (E120 and Q123) that coordinate Ca(2+) binding being conserved. In these previous studies, while the tetramer crystallized at neutral pH, the pentamer crystallized at low pH, suggesting that preference for a particular oligomeric state is pH dependent and that pH could influence Ca(2+) binding. Here, we sought to examine if the CCD of NSP4 from a single RV strain can exist in two oligomeric states regulated by Ca(2+) or pH. Biochemical, biophysical, and crystallographic studies show that while the CCD of SA11 NSP4 exhibits high-affinity binding to Ca(2+) at neutral pH and forms a tetramer, it does not bind Ca(2+) at low pH and forms a pentamer, and the transition from tetramer to pentamer is reversible with pH. Mutational analysis shows that Ca(2+) binding is necessary for the tetramer formation, as an E120A mutant forms a pentamer. We propose that the structural plasticity of NSP4 regulated by pH and Ca(2+) may form a basis for its pleiotropic functions during RV replication.
Importance: The nonstructural protein NSP4 of rotavirus is a multifunctional protein that plays an important role in virus replication, morphogenesis, and pathogenesis. Previous crystallography studies of the coiled-coil domain (CCD) of NSP4 from two different rotavirus strains showed two distinct oligomeric states, a Ca(2+)-bound tetrameric state and a Ca(2+)-free pentameric state. Whether NSP4 CCD from the same strain can exist in different oligomeric states and what factors might regulate its oligomeric preferences are not known. This study used a combination of biochemical, biophysical, and crystallography techniques and found that the NSP4 CCD can undergo a reversible transition from a Ca(2+)-bound tetramer to a Ca(2+)-free pentamer in response to changes in pH. From these studies, we hypothesize that this remarkable structural adaptability of the CCD forms a basis for the pleiotropic functional properties of NSP4.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
New tetrameric forms of the rotavirus NSP4 with antiparallel helices.Arch Virol. 2018 Jun;163(6):1531-1547. doi: 10.1007/s00705-018-3753-6. Epub 2018 Feb 17. Arch Virol. 2018. PMID: 29455326
-
Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4.J Virol. 2011 Dec;85(23):12721-32. doi: 10.1128/JVI.00349-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917949 Free PMC article.
-
Mutational analysis of the rotavirus NSP4 enterotoxic domain that binds to caveolin-1.Virol J. 2013 Nov 13;10:336. doi: 10.1186/1743-422X-10-336. Virol J. 2013. PMID: 24220211 Free PMC article.
-
Rotavirus protein expression is important for virus assembly and pathogenesis.Arch Virol Suppl. 1996;12:69-77. doi: 10.1007/978-3-7091-6553-9_8. Arch Virol Suppl. 1996. PMID: 9015103 Review.
-
Rotavirus NSP4: a multifunctional viral enterotoxin.Viral Immunol. 2005;18(1):27-40. doi: 10.1089/vim.2005.18.27. Viral Immunol. 2005. PMID: 15802952 Review. No abstract available.
Cited by
-
Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology.J Virol. 2015 Oct 7;90(1):43-56. doi: 10.1128/JVI.01930-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26446608 Free PMC article.
-
Rotavirus induces intercellular calcium waves through ADP signaling.Science. 2020 Nov 20;370(6519):eabc3621. doi: 10.1126/science.abc3621. Science. 2020. PMID: 33214249 Free PMC article.
-
Effect of Formulation Variables on the Stability of a Live, Rotavirus (RV3-BB) Vaccine Candidate using in vitro Gastric Digestion Models to Mimic Oral Delivery.J Pharm Sci. 2021 Feb;110(2):760-770. doi: 10.1016/j.xphs.2020.09.047. Epub 2020 Oct 7. J Pharm Sci. 2021. PMID: 33035539 Free PMC article.
-
The Emerging Roles of Viroporins in ER Stress Response and Autophagy Induction during Virus Infection.Viruses. 2015 Jun 4;7(6):2834-57. doi: 10.3390/v7062749. Viruses. 2015. PMID: 26053926 Free PMC article. Review.
-
Enterotoxigenic Escherichia coli Heat-Stable Toxin and Ebola Virus Delta Peptide: Similarities and Differences.Pathogens. 2022 Jan 27;11(2):170. doi: 10.3390/pathogens11020170. Pathogens. 2022. PMID: 35215114 Free PMC article. Review.
References
-
- Bhowmick R, Halder UC, Chattopadhyay S, Chanda S, Nandi S, Bagchi P, Nayak MK, Chakrabarti O, Kobayashi N, Chawla-Sarkar M. 2012. Rotaviral enterotoxin nonstructural protein 4 targets mitochondria for activation of apoptosis during infection. J. Biol. Chem. 287:35004–35020. 10.1074/jbc.M112.369595. - DOI - PMC - PubMed
-
- Chattopadhyay S, Basak T, Nayak MK, Bhardwaj G, Mukherjee A, Bhowmick R, Sengupta S, Chakrabarti O, Chatterjee NS, Chawla-Sarkar M. 2013. Identification of cellular calcium binding protein calmodulin as a regulator of rotavirus A infection during comparative proteomic study. PLoS One 8:e56655. 10.1371/journal.pone.0056655. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

