Members of the human gut microbiota involved in recovery from Vibrio cholerae infection
- PMID: 25231861
- PMCID: PMC4353411
- DOI: 10.1038/nature13738
Members of the human gut microbiota involved in recovery from Vibrio cholerae infection
Abstract
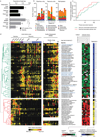
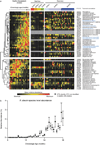
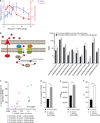
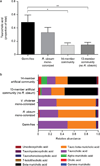
Given the global burden of diarrhoeal diseases, it is important to understand how members of the gut microbiota affect the risk for, course of, and recovery from disease in children and adults. The acute, voluminous diarrhoea caused by Vibrio cholerae represents a dramatic example of enteropathogen invasion and gut microbial community disruption. Here we conduct a detailed time-series metagenomic study of faecal microbiota collected during the acute diarrhoeal and recovery phases of cholera in a cohort of Bangladeshi adults living in an area with a high burden of disease. We find that recovery is characterized by a pattern of accumulation of bacterial taxa that shows similarities to the pattern of assembly/maturation of the gut microbiota in healthy Bangladeshi children. To define the underlying mechanisms, we introduce into gnotobiotic mice an artificial community composed of human gut bacterial species that directly correlate with recovery from cholera in adults and are indicative of normal microbiota maturation in healthy Bangladeshi children. One of the species, Ruminococcus obeum, exhibits consistent increases in its relative abundance upon V. cholerae infection of the mice. Follow-up analyses, including mono- and co-colonization studies, establish that R. obeum restricts V. cholerae colonization, that R. obeum luxS (autoinducer-2 (AI-2) synthase) expression and AI-2 production increase significantly with V. cholerae invasion, and that R. obeum AI-2 causes quorum-sensing-mediated repression of several V. cholerae colonization factors. Co-colonization with V. cholerae mutants discloses that R. obeum AI-2 reduces Vibrio colonization/pathogenicity through a novel pathway that does not depend on the V. cholerae AI-2 sensor, LuxP. The approach described can be used to mine the gut microbiota of Bangladeshi or other populations for members that use autoinducers and/or other mechanisms to limit colonization with V. cholerae, or conceivably other enteropathogens.
Figures







Comment in
-
Bacterial pathogenesis: First aid kit for cholera.Nat Rev Microbiol. 2014 Nov;12(11):726. doi: 10.1038/nrmicro3366. Epub 2014 Sep 29. Nat Rev Microbiol. 2014. PMID: 25263221 No abstract available.
-
Can chatter between microbes prevent cholera?Trends Microbiol. 2014 Dec;22(12):660-2. doi: 10.1016/j.tim.2014.10.006. Epub 2014 Nov 4. Trends Microbiol. 2014. PMID: 25468792
-
Microbiota talks cholera out of the gut.Cell Host Microbe. 2014 Nov 12;16(5):549-50. doi: 10.1016/j.chom.2014.10.011. Epub 2014 Nov 12. Cell Host Microbe. 2014. PMID: 25525783
References
-
- WHO. Cholera, 2011. Weekly Epidemiological Record. 2012;87:289–304. - PubMed
-
- Dufrene M, Legendre P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monographs. 1997;67:345–366.
REFERENCES FOR ONLINE METHODS AND SUPPLEMENTARY INFORMATION
-
- Faiz MA, Basher A. Antimicrobial resistance: Bangladesh experience. Regional Health Forum. 2011;15:1–18.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

