Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland
- PMID: 25232158
- PMCID: PMC4313278
- DOI: 10.1128/JCM.01959-14
Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland
Abstract
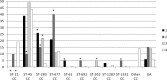
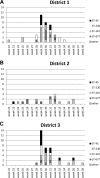
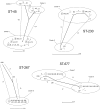
A total of 95 human Campylobacter jejuni isolates acquired from domestic infections and collected from three districts in Finland during the seasonal peak (June to September) in 2012 were analyzed by PCR-based multilocus sequence typing (MLST) and by whole-genome sequencing (WGS). Four predominant sequence types (STs) were detected among the isolates: ST-45 (21%) and ST-230 (14%, ST-45 clonal complex [CC]), ST-267 (21%, ST-283 CC), and ST-677 (19%, ST-677 CC). In districts 1 and 3, most of the infections occurred from early July to the middle of August, with a peak at weeks 29 to 31, but in district 2, the infections were dispersed more evenly throughout 3 months (June to August). WGS data were used for further whole-genome MLST (wgMLST) analyses of the isolates representing the four common STs. Shared loci of the isolates within each ST were analyzed as distance matrices of allelic profiles by the neighbor-net algorithm. The highest allelic variations (>400 different alleles) were detected between different clusters of ST-45 isolates (1,121 shared loci), while ST-230 (1,264 shared loci), ST-677 (1,169 shared loci), and ST-267 isolates (1,217 shared loci) were less diverse with the clusters differing by <40 alleles. Closely related isolates showing no allelic variation (subclusters) were detected among all four major STs. In some cases, they originated from different districts, suggesting that isolates can be epidemiologically connected and may have the same infection source despite being originally identified as sporadic infections.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Tracing isolates from domestic human Campylobacter jejuni infections to chicken slaughter batches and swimming water using whole-genome multilocus sequence typing.Int J Food Microbiol. 2016 Jun 2;226:53-60. doi: 10.1016/j.ijfoodmicro.2016.03.009. Epub 2016 Mar 20. Int J Food Microbiol. 2016. PMID: 27041390
-
Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland.Appl Environ Microbiol. 2010 Aug;76(15):5228-36. doi: 10.1128/AEM.00581-10. Epub 2010 Jun 11. Appl Environ Microbiol. 2010. PMID: 20543048 Free PMC article.
-
Population Genetics and Characterization of Campylobacter jejuni Isolates from Western Jackdaws and Game Birds in Finland.Appl Environ Microbiol. 2019 Feb 6;85(4):e02365-18. doi: 10.1128/AEM.02365-18. Print 2019 Feb 15. Appl Environ Microbiol. 2019. PMID: 30552190 Free PMC article.
-
A combination of MLST and CRISPR typing reveals dominant Campylobacter jejuni types in organically farmed laying hens.J Appl Microbiol. 2014 Jul;117(1):249-57. doi: 10.1111/jam.12503. Epub 2014 Apr 8. J Appl Microbiol. 2014. PMID: 24655229
-
Epidemiological relationships of Campylobacter jejuni strains isolated from humans and chickens in South Korea.J Microbiol. 2017 Jan;55(1):13-20. doi: 10.1007/s12275-017-6308-8. Epub 2016 Dec 30. J Microbiol. 2017. PMID: 28035601
Cited by
-
Whole-Genome Sequencing to Detect Numerous Campylobacter jejuni Outbreaks and Match Patient Isolates to Sources, Denmark, 2015-2017.Emerg Infect Dis. 2020 Mar;26(3):523-532. doi: 10.3201/eid2603.190947. Emerg Infect Dis. 2020. PMID: 32091364 Free PMC article.
-
Molecular Assay Validation Using Genomic Sequence Databases.J Clin Microbiol. 2016 Dec;54(12):2854-2856. doi: 10.1128/JCM.01797-16. Epub 2016 Oct 12. J Clin Microbiol. 2016. PMID: 27733633 Free PMC article.
-
Molecular Detection and Epidemiological Features of Selected Bacterial, Viral, and Parasitic Enteropathogens in Stool Specimens from Children with Acute Diarrhea in Thi-Qar Governorate, Iraq.Int J Environ Res Public Health. 2019 May 6;16(9):1573. doi: 10.3390/ijerph16091573. Int J Environ Res Public Health. 2019. PMID: 31064051 Free PMC article.
-
Whole-Genome Sequencing in Epidemiology of Campylobacter jejuni Infections.J Clin Microbiol. 2017 May;55(5):1269-1275. doi: 10.1128/JCM.00017-17. Epub 2017 Mar 1. J Clin Microbiol. 2017. PMID: 28249998 Free PMC article. Review.
-
Clinical impact of major pathogenic genotypes of Pseudomonas aeruginosa associated with refractory chronic suppurative otitis media.Eur J Clin Microbiol Infect Dis. 2024 Dec;43(12):2429-2440. doi: 10.1007/s10096-024-04957-x. Epub 2024 Oct 14. Eur J Clin Microbiol Infect Dis. 2024. PMID: 39400676
References
-
- World Health Organization. 2012. The global view of campylobacteriosis: report of an expert consultation. World Health Organization, Utrecht, Netherlands.
-
- Nylen G, Dunstan F, Palmer SR, Andersson Y, Bager F, Cowden J, Feierl G, Galloway Y, Kapperud G, Megraud F, Molbak K, Petersen LR, Ruutu P. 2002. The seasonal distribution of Campylobacter infection in nine European countries and New Zealand. Epidemiol. Infect. 128:383–390. 10.1017/S0950268802006830. - DOI - PMC - PubMed
-
- de Haan CPA, Kivistö RI, Rautelin H, Hänninen M. 2014. How molecular typing has changed our understanding on sources and transmission routes of campylobacteriosis in Finland, p 241 In Sheppard SK. (ed), Campylobacter ecology and evolution. Caister Academic Press, Swansea, United Kingdom.
-
- Jore S, Viljugrein H, Brun E, Heier BT, Borck B, Ethelberg S, Hakkinen M, Kuusi M, Reiersen J, Hansson I, Engvall EO, Lofdahl M, Wagenaar JA, van Pelt W, Hofshagen M. 2010. Trends in Campylobacter incidence in broilers and humans in six European countries, 1997-2007. Prev. Vet. Med. 93:33–41. 10.1016/j.prevetmed.2009.09.015. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

