Out of balance: R-loops in human disease
- PMID: 25233079
- PMCID: PMC4169248
- DOI: 10.1371/journal.pgen.1004630
Out of balance: R-loops in human disease
Abstract
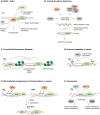
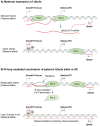
R-loops are cellular structures composed of an RNA/DNA hybrid, which is formed when the RNA hybridises to a complementary DNA strand and a displaced single-stranded DNA. R-loops have been detected in various organisms from bacteria to mammals and play crucial roles in regulating gene expression, DNA and histone modifications, immunoglobulin class switch recombination, DNA replication, and genome stability. Recent evidence suggests that R-loops are also involved in molecular mechanisms of neurological diseases and cancer. In addition, mutations in factors implicated in R-loop biology, such as RNase H and SETX (senataxin), lead to devastating human neurodegenerative disorders, highlighting the importance of correctly regulating the level of R-loops in human cells. In this review we summarise current advances in this field, with a particular focus on diseases associated with dysregulation of R-loop structures. We also discuss potential therapeutic approaches for such diseases and highlight future research directions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- White RL, Hogness DS (1977) R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell 10: 177–192. - PubMed
-
- Drolet M, Bi X, Liu LF (1994) Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem 269: 2068–2074. - PubMed
-
- Aguilera A, Garcia-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

