NESmapper: accurate prediction of leucine-rich nuclear export signals using activity-based profiles
- PMID: 25233087
- PMCID: PMC4168985
- DOI: 10.1371/journal.pcbi.1003841
NESmapper: accurate prediction of leucine-rich nuclear export signals using activity-based profiles
Abstract
The nuclear export of proteins is regulated largely through the exportin/CRM1 pathway, which involves the specific recognition of leucine-rich nuclear export signals (NESs) in the cargo proteins, and modulates nuclear-cytoplasmic protein shuttling by antagonizing the nuclear import activity mediated by importins and the nuclear import signal (NLS). Although the prediction of NESs can help to define proteins that undergo regulated nuclear export, current methods of predicting NESs, including computational tools and consensus-sequence-based searches, have limited accuracy, especially in terms of their specificity. We found that each residue within an NES largely contributes independently and additively to the entire nuclear export activity. We created activity-based profiles of all classes of NESs with a comprehensive mutational analysis in mammalian cells. The profiles highlight a number of specific activity-affecting residues not only at the conserved hydrophobic positions but also in the linker and flanking regions. We then developed a computational tool, NESmapper, to predict NESs by using profiles that had been further optimized by training and combining the amino acid properties of the NES-flanking regions. This tool successfully reduced the considerable number of false positives, and the overall prediction accuracy was higher than that of other methods, including NESsential and Wregex. This profile-based prediction strategy is a reliable way to identify functional protein motifs. NESmapper is available at http://sourceforge.net/projects/nesmapper.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

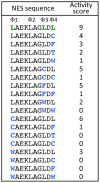


Similar articles
-
Sequence and structural analyses of nuclear export signals in the NESdb database.Mol Biol Cell. 2012 Sep;23(18):3677-93. doi: 10.1091/mbc.E12-01-0046. Epub 2012 Jul 25. Mol Biol Cell. 2012. PMID: 22833565 Free PMC article.
-
Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals.Proc Natl Acad Sci U S A. 1999 May 25;96(11):6229-34. doi: 10.1073/pnas.96.11.6229. Proc Natl Acad Sci U S A. 1999. PMID: 10339570 Free PMC article.
-
Prediction of nuclear export signals using weighted regular expressions (Wregex).Bioinformatics. 2014 May 1;30(9):1220-7. doi: 10.1093/bioinformatics/btu016. Epub 2014 Jan 9. Bioinformatics. 2014. PMID: 24413524
-
Leucine-rich nuclear-export signals: born to be weak.Trends Cell Biol. 2005 Mar;15(3):121-4. doi: 10.1016/j.tcb.2005.01.005. Trends Cell Biol. 2005. PMID: 15752974 Review.
-
Recognition of nuclear targeting signals by Karyopherin-β proteins.Curr Opin Struct Biol. 2010 Dec;20(6):782-90. doi: 10.1016/j.sbi.2010.09.008. Epub 2010 Oct 13. Curr Opin Struct Biol. 2010. PMID: 20951026 Free PMC article. Review.
Cited by
-
Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein.J Virol. 2015 Mar;89(6):3038-48. doi: 10.1128/JVI.03125-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552709 Free PMC article.
-
Target gene responses differ when transcription factor levels are acutely decreased by nuclear export versus degradation.bioRxiv [Preprint]. 2024 May 20:2024.05.20.595009. doi: 10.1101/2024.05.20.595009. bioRxiv. 2024. Update in: Development. 2024 Nov 1;151(21):dev202775. doi: 10.1242/dev.202775. PMID: 38826476 Free PMC article. Updated. Preprint.
-
Proteome-wide search for functional motifs altered in tumors: Prediction of nuclear export signals inactivated by cancer-related mutations.Sci Rep. 2016 May 12;6:25869. doi: 10.1038/srep25869. Sci Rep. 2016. PMID: 27174732 Free PMC article.
-
Description of USP12 nuclear export sequence.Proc Natl Acad Sci U S A. 2016 Jun 14;113(24):E3315-6. doi: 10.1073/pnas.1606081113. Epub 2016 Jun 2. Proc Natl Acad Sci U S A. 2016. PMID: 27302950 Free PMC article. No abstract available.
-
Alternative splicing of HDAC7 regulates its interaction with 14-3-3 proteins to alter histone marks and target gene expression.Cell Rep. 2023 Mar 28;42(3):112273. doi: 10.1016/j.celrep.2023.112273. Epub 2023 Mar 17. Cell Rep. 2023. PMID: 36933216 Free PMC article.
References
-
- Ossareh-Nazari B, Gwizdek C, Dargemont C (2001) Protein export from the nucleus. Traffic 2: 684–689. - PubMed
-
- Fornerod M, Ohno M (2002) Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl Cell Differ 35: 67–91. - PubMed
-
- Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

