Crimean-Congo hemorrhagic fever virus entry into host cells occurs through the multivesicular body and requires ESCRT regulators
- PMID: 25233119
- PMCID: PMC4169490
- DOI: 10.1371/journal.ppat.1004390
Crimean-Congo hemorrhagic fever virus entry into host cells occurs through the multivesicular body and requires ESCRT regulators
Abstract
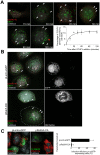
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne bunyavirus causing outbreaks of severe disease in humans, with a fatality rate approaching 30%. There are no widely accepted therapeutics available to prevent or treat the disease. CCHFV enters host cells through clathrin-mediated endocytosis and is subsequently transported to an acidified compartment where the fusion of virus envelope with cellular membranes takes place. To better understand the uptake pathway, we sought to identify host factors controlling CCHFV transport through the cell. We demonstrate that after passing through early endosomes in a Rab5-dependent manner, CCHFV is delivered to multivesicular bodies (MVBs). Virus particles localized to MVBs approximately 1 hour after infection and affected the distribution of the organelle within cells. Interestingly, blocking Rab7 activity had no effect on association of the virus with MVBs. Productive virus infection depended on phosphatidylinositol 3-kinase (PI3K) activity, which meditates the formation of functional MVBs. Silencing Tsg101, Vps24, Vps4B, or Alix/Aip1, components of the endosomal sorting complex required for transport (ESCRT) pathway controlling MVB biogenesis, inhibited infection of wild-type virus as well as a novel pseudotyped vesicular stomatitis virus (VSV) bearing CCHFV glycoprotein, supporting a role for the MVB pathway in CCHFV entry. We further demonstrate that blocking transport out of MVBs still allowed virus entry while preventing vesicular acidification, required for membrane fusion, trapped virions in the MVBs. These findings suggest that MVBs are necessary for infection and are the sites of virus-endosome membrane fusion.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Old world arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport.PLoS Pathog. 2011 Sep;7(9):e1002232. doi: 10.1371/journal.ppat.1002232. Epub 2011 Sep 8. PLoS Pathog. 2011. PMID: 21931550 Free PMC article.
-
Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway.Virology. 2013 Sep;444(1-2):45-54. doi: 10.1016/j.virol.2013.05.030. Epub 2013 Jun 19. Virology. 2013. PMID: 23791227
-
LDLR is an entry receptor for Crimean-Congo hemorrhagic fever virus.Cell Res. 2024 Feb;34(2):140-150. doi: 10.1038/s41422-023-00917-w. Epub 2024 Jan 5. Cell Res. 2024. PMID: 38182887 Free PMC article.
-
Crimean-Congo Hemorrhagic Fever: Tick-Host-Virus Interactions.Front Cell Infect Microbiol. 2017 May 26;7:213. doi: 10.3389/fcimb.2017.00213. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28603698 Free PMC article. Review.
-
The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review.
Cited by
-
Orthobunyaviruses: From Virus Binding to Penetration into Mammalian Host Cells.Viruses. 2021 May 10;13(5):872. doi: 10.3390/v13050872. Viruses. 2021. PMID: 34068494 Free PMC article. Review.
-
Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus.J Biol Chem. 2018 Jun 29;293(26):9937-9944. doi: 10.1074/jbc.RA118.002494. Epub 2018 Apr 20. J Biol Chem. 2018. PMID: 29678879 Free PMC article.
-
KSHV Entry and Trafficking in Target Cells-Hijacking of Cell Signal Pathways, Actin and Membrane Dynamics.Viruses. 2016 Nov 14;8(11):305. doi: 10.3390/v8110305. Viruses. 2016. PMID: 27854239 Free PMC article. Review.
-
ESCRT-I Protein Tsg101 Plays a Role in the Post-macropinocytic Trafficking and Infection of Endothelial Cells by Kaposi's Sarcoma-Associated Herpesvirus.PLoS Pathog. 2016 Oct 20;12(10):e1005960. doi: 10.1371/journal.ppat.1005960. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27764233 Free PMC article.
-
ESCRT puts its thumb on the nanoscale: Fixing tiny holes in endolysosomes.Curr Opin Cell Biol. 2020 Aug;65:122-130. doi: 10.1016/j.ceb.2020.06.002. Epub 2020 Jul 27. Curr Opin Cell Biol. 2020. PMID: 32731154 Free PMC article. Review.
References
-
- Estrada-Pena A, Ruiz-Fons F, Acevedo P, Gortazar C, de la Fuente J (2013) Factors driving the circulation and possible expansion of Crimean-Congo haemorrhagic fever virus in the western Palearctic. J Appl Microbiol 114: 278–286. - PubMed
-
- Whitehouse CA (2004) Crimean-Congo hemorrhagic fever. Antiviral Res 64: 145–160. - PubMed
-
- Vorou R, Pierroutsakos IN, Maltezou HC (2007) Crimean-Congo hemorrhagic fever. Curr Opin Infect Dis 20: 495–500. - PubMed
-
- Schmaljohn CS, Nichol S T. (2007) Bunyaviridae. In: Knipe DM, Howley P M., editor. Fields Virology. Philadelphia: Lippincott Williams & Wilkins. pp. 1741–1789.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

