An optimized optogenetic clustering tool for probing protein interaction and function
- PMID: 25233328
- PMCID: PMC4170572
- DOI: 10.1038/ncomms5925
An optimized optogenetic clustering tool for probing protein interaction and function
Abstract
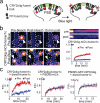
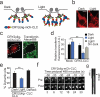
The Arabidopsis photoreceptor cryptochrome 2 (CRY2) was previously used as an optogenetic module, allowing spatiotemporal control of cellular processes with light. Here we report the development of a new CRY2-derived optogenetic module, 'CRY2olig', which induces rapid, robust, and reversible protein oligomerization in response to light. Using this module, we developed a novel protein interaction assay, Light-Induced Co-clustering, that can be used to interrogate protein interaction dynamics in live cells. In addition to use probing protein interactions, CRY2olig can also be used to induce and reversibly control diverse cellular processes with spatial and temporal resolution. Here we demonstrate disrupting clathrin-mediated endocytosis and promoting Arp2/3-mediated actin polymerization with light. These new CRY2-based approaches expand the growing arsenal of optogenetic strategies to probe cellular function.
Figures



Comment in
-
Cell biology: Versatile clusters formed by light.Nat Methods. 2014 Nov;11(11):1087. doi: 10.1038/nmeth.3161. Nat Methods. 2014. PMID: 25551121 No abstract available.
References
-
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. - PubMed
-
- Petschnigg J, Snider J, Stagljar I. Interactive proteomics research technologies: recent applications and advances. Curr. Opin. Biotechnol. 2011;22:50–58. - PubMed
-
- Buskirk AR, Liu DR. Creating Small-Molecule-Dependent Switches to Modulate Biological Functions. Chem. Biol. 2005;12:151–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

