OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway
- PMID: 25233933
- PMCID: PMC4176851
- DOI: 10.1186/1476-4598-13-218
OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway
Abstract
Background: MicroRNA-196 (miR-196), which is highly up-regulated in oral cancer cells, has been reported to be aberrantly expressed in several cancers; however, the significance of miR-196 in oral cancer has not yet been addressed.
Methods: Cellular functions in response to miR-196 modulation were examined, including cell growth, migration, invasion and radio/chemosensitivity. Algorithm-based studies were used to identify the regulatory target of miR-196. The miR-196 target gene and downstream molecular mechanisms were confirmed by RT-qPCR, western blot, luciferase reporter and confocal microscopy analyses. miR-196 expression was determined in paired cancer and adjacent normal tissues from oral cancer patients.
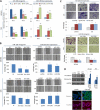
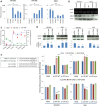
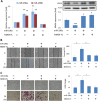
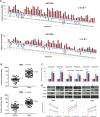
Results: Both miR-196a and miR-196b were highly over-expressed in the cancer tissue and correlated with lymph node metastasis (P = 0.001 and P = 0.006, respectively). Functionally, miR-196 actively promoted cell migration and invasion without affecting cell growth. Mechanistically, miR-196 performed it's their function by inhibiting NME4 expression and further activating p-JNK, suppressing TIMP1, and augmenting MMP1/9.
Conclusion: miR-196 contributes to oral cancer by promoting cell migration and invasion. Clinically, miR-196a/b was significantly over-expressed in the cancer tissues and correlated with lymph node metastasis. Thus, our findings provide new knowledge of the underlying mechanism of cancer metastasis. miR-196 may serve as a promising marker for better oral cancer management.
Figures

References
-
- Stucken E, Weissman J, Spiegel JH. Oral cavity risk factors: experts' opinions and literature support. J Otolaryngol. 2010;39:76–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

