Single nucleotide variations: biological impact and theoretical interpretation
- PMID: 25234433
- PMCID: PMC4253807
- DOI: 10.1002/pro.2552
Single nucleotide variations: biological impact and theoretical interpretation
Abstract
Genome-wide association studies (GWAS) and whole-exome sequencing (WES) generate massive amounts of genomic variant information, and a major challenge is to identify which variations drive disease or contribute to phenotypic traits. Because the majority of known disease-causing mutations are exonic non-synonymous single nucleotide variations (nsSNVs), most studies focus on whether these nsSNVs affect protein function. Computational studies show that the impact of nsSNVs on protein function reflects sequence homology and structural information and predict the impact through statistical methods, machine learning techniques, or models of protein evolution. Here, we review impact prediction methods and discuss their underlying principles, their advantages and limitations, and how they compare to and complement one another. Finally, we present current applications and future directions for these methods in biological research and medical genetics.
Keywords: disease causing SNV (single nucleotide variation); functional impact prediction methods; missense variant classification; non-synonymous protein mutations; single nucleotide polymorphism prioritization.
© 2014 The Authors Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures
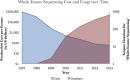



References
-
- Scopus—document search. Available at: < http://www.scopus.com/ >. Accessed 2014.
-
- DNA Sequencing Costs. Available at: < http://www.genome.gov/sequencingcosts/ >. Accessed on June 15, 2014.
-
- Whole exome seq—compare prices and order services—science exchange. Available at: < https://www.scienceexchange.com/services/whole-exome- seq >. Accessed on June 24, 2014.
-
- Hayden EC. Technology: the $1,000 genome. Nature. 2014;507:294–295. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

