Targeting environmental adaptation in the monocot model Brachypodium distachyon: a multi-faceted approach
- PMID: 25236859
- PMCID: PMC4177692
- DOI: 10.1186/1471-2164-15-801
Targeting environmental adaptation in the monocot model Brachypodium distachyon: a multi-faceted approach
Abstract
Background: The local environment plays a major role in the spatial distribution of plant populations. Natural plant populations have an extremely poor displacing capacity, so their continued survival in a given environment depends on how well they adapt to local pedoclimatic conditions. Genomic tools can be used to identify adaptive traits at a DNA level and to further our understanding of evolutionary processes. Here we report the use of genotyping-by-sequencing on local groups of the sequenced monocot model species Brachypodium distachyon. Exploiting population genetics, landscape genomics and genome wide association studies, we evaluate B. distachyon role as a natural probe for identifying genomic loci involved in environmental adaptation.
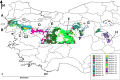
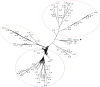
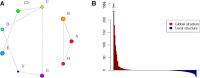
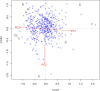
Results: Brachypodium distachyon individuals were sampled in nine locations with different ecologies and characterized with 16,697 SNPs. Variations in sequencing depth showed consistent patterns at 8,072 genomic bins, which were significantly enriched in transposable elements. We investigated the structuration and diversity of this collection, and exploited climatic data to identify loci with adaptive significance through i) two different approaches for genome wide association analyses considering climatic variation, ii) an outlier loci approach, and iii) a canonical correlation analysis on differentially sequenced bins. A linkage disequilibrium-corrected Bonferroni method was applied to filter associations. The two association methods jointly identified a set of 15 genes significantly related to environmental adaptation. The outlier loci approach revealed that 5.7% of the loci analysed were under selection. The canonical correlation analysis showed that the distribution of some differentially sequenced regions was associated to environmental variation.
Conclusions: We show that the multi-faceted approach used here targeted different components of B. distachyon adaptive variation, and may lead to the discovery of genes related to environmental adaptation in natural populations. Its application to a model species with a fully sequenced genome is a modular strategy that enables the stratification of biological material and thus improves our knowledge of the functional loci determining adaptation in near-crop species. When coupled with population genetics and measures of genomic structuration, methods coming from genome wide association studies may lead to the exploitation of model species as natural probes to identify loci related to environmental adaptation.
Figures



Similar articles
-
Genome-wide scans of selection highlight the impact of biotic and abiotic constraints in natural populations of the model grass Brachypodium distachyon.Plant J. 2018 Oct;96(2):438-451. doi: 10.1111/tpj.14042. Epub 2018 Sep 1. Plant J. 2018. PMID: 30044522
-
The evolution of transposable elements in Brachypodium distachyon is governed by purifying selection, while neutral and adaptive processes play a minor role.Elife. 2024 Apr 12;12:RP93284. doi: 10.7554/eLife.93284. Elife. 2024. PMID: 38606833 Free PMC article.
-
Molecular diversity and landscape genomics of the crop wild relative Triticum urartu across the Fertile Crescent.Plant J. 2018 May;94(4):670-684. doi: 10.1111/tpj.13888. Epub 2018 Apr 18. Plant J. 2018. PMID: 29573496
-
The search for loci under selection: trends, biases and progress.Mol Ecol. 2018 Mar;27(6):1342-1356. doi: 10.1111/mec.14549. Epub 2018 Mar 30. Mol Ecol. 2018. PMID: 29524276 Review.
-
Brachypodium: A Monocot Grass Model Genus for Plant Biology.Plant Cell. 2018 Aug;30(8):1673-1694. doi: 10.1105/tpc.18.00083. Epub 2018 Jul 11. Plant Cell. 2018. PMID: 29997238 Free PMC article. Review.
Cited by
-
Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions.Am Nat. 2016 Oct;188(4):379-97. doi: 10.1086/688018. Epub 2016 Aug 15. Am Nat. 2016. PMID: 27622873 Free PMC article. Review.
-
Population genomics and climate adaptation of a C4 perennial grass, Panicum hallii (Poaceae).BMC Genomics. 2018 Nov 1;19(1):792. doi: 10.1186/s12864-018-5179-7. BMC Genomics. 2018. PMID: 30384830 Free PMC article.
-
Genetic and Methylome Variation in Turkish Brachypodium Distachyon Accessions Differentiate Two Geographically Distinct Subpopulations.Int J Mol Sci. 2020 Sep 13;21(18):6700. doi: 10.3390/ijms21186700. Int J Mol Sci. 2020. PMID: 32933168 Free PMC article.
-
Never the Two Shall Mix: Robust Indel Markers to Ensure the Fidelity of Two Pivotal and Closely-Related Accessions of Brachypodium distachyon.Plants (Basel). 2019 Jun 6;8(6):153. doi: 10.3390/plants8060153. Plants (Basel). 2019. PMID: 31174296 Free PMC article.
-
Migration without interbreeding: Evolutionary history of a highly selfing Mediterranean grass inferred from whole genomes.Mol Ecol. 2022 Jan;31(1):70-85. doi: 10.1111/mec.16207. Epub 2021 Oct 17. Mol Ecol. 2022. PMID: 34601787 Free PMC article.
References
-
- Hughes AL. Adaptive Evolution of Genes and Genomes. New York: Oxford University Press; 1999.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

