Exploring metabolic pathways and regulation through functional chemoproteomic and metabolomic platforms
- PMID: 25237861
- PMCID: PMC4171689
- DOI: 10.1016/j.chembiol.2014.07.007
Exploring metabolic pathways and regulation through functional chemoproteomic and metabolomic platforms
Abstract
Genome sequencing efforts have revealed a strikingly large number of uncharacterized genes, including poorly or uncharacterized metabolic enzymes, metabolites, and metabolic networks that operate in normal physiology, and those enzymes and pathways that may be rewired under pathological conditions. Although deciphering the functions of the uncharacterized metabolic genome is a challenging prospect, it also presents an opportunity for identifying novel metabolic nodes that may be important in disease therapy. In this review, we will discuss the chemoproteomic and metabolomic platforms used in identifying, characterizing, and targeting nodal metabolic pathways important in physiology and disease, describing an integrated workflow for functional mapping of metabolic enzymes.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
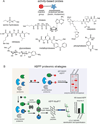
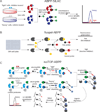


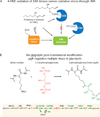

References
-
- Adam GC, Cravatt BF, Sorensen EJ. Profiling the specific reactivity of the proteome with non-directed activity-based probes. Chem. Biol. 2001;8:81–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

