Mining the metabiome: identifying novel natural products from microbial communities
- PMID: 25237864
- PMCID: PMC4171686
- DOI: 10.1016/j.chembiol.2014.08.006
Mining the metabiome: identifying novel natural products from microbial communities
Abstract
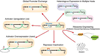
Microbial-derived natural products provide the foundation for most of the chemotherapeutic arsenal available to contemporary medicine. In the face of a dwindling pipeline of new lead structures identified by traditional culturing techniques and an increasing need for new therapeutics, surveys of microbial biosynthetic diversity across environmental metabiomes have revealed enormous reservoirs of as yet untapped natural products chemistry. In this review, we touch on the historical context of microbial natural product discovery and discuss innovations and technological advances that are facilitating culture-dependent and culture-independent access to new chemistry from environmental microbiomes with the goal of reinvigorating the small molecule therapeutics discovery pipeline. We highlight the successful strategies that have emerged and some of the challenges that must be overcome to enable the development of high-throughput methods for natural product discovery from complex microbial communities.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

References
-
- Aakvik T, Degnes KF, Dahlsrud R, Schmidt F, Dam R, Yu L, Völker U, Ellingsen TE, Valla S. A plasmid RK2-based broad-host-range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol. Lett. 2009;296:149–158. - PubMed
-
- Baker DD, Chu M, Oza U, Rajgarhia V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007;24:1225–1244. - PubMed
-
- Baker M. Gene editing at CRISPR speed. Nat. Biotechnol. 2014;32:309–312. - PubMed
-
- Ben-Dov E, Kramarsky-Winter E, Kushmaro A. An in situ method for cultivating microorganisms using a double encapsulation technique. FEMS Microbiol. Ecol. 2009;68:363–371. - PubMed
-
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

