DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm
- PMID: 25238154
- PMCID: PMC4169571
- DOI: 10.1371/journal.pone.0107653
DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm
Abstract
Background: Prenatal maternal stress (PNMS) predicts a wide variety of behavioral and physical outcomes in the offspring. Although epigenetic processes may be responsible for PNMS effects, human research is hampered by the lack of experimental methods that parallel controlled animal studies. Disasters, however, provide natural experiments that can provide models of prenatal stress.
Methods: Five months after the 1998 Quebec ice storm we recruited women who had been pregnant during the disaster and assessed their degrees of objective hardship and subjective distress. Thirteen years later, we investigated DNA methylation profiling in T cells obtained from 36 of the children, and compared selected results with those from saliva samples obtained from the same children at age 8.
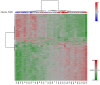
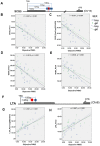
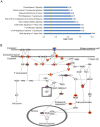
Results: Prenatal maternal objective hardship was correlated with DNA methylation levels in 1675 CGs affiliated with 957 genes predominantly related to immune function; maternal subjective distress was uncorrelated. DNA methylation changes in SCG5 and LTA, both highly correlated with maternal objective stress, were comparable in T cells, peripheral blood mononuclear cells (PBMCs) and saliva cells.
Conclusions: These data provide first evidence in humans supporting the conclusion that PNMS results in a lasting, broad, and functionally organized DNA methylation signature in several tissues in offspring. By using a natural disaster model, we can infer that the epigenetic effects found in Project Ice Storm are due to objective levels of hardship experienced by the pregnant woman rather than to her level of sustained distress.
Conflict of interest statement
Figures

References
-
- Weinstock M (2008) The long-term behavioural consequences of prenatal stress. Neuroscience and Biobehavioral Reviews 32: 1073–1086. - PubMed
-
- Charil A, Laplante DP, Vaillancourt C, King S (2010) Prenatal stress and brain development. Brain Res Rev 65: 56–79. - PubMed
-
- Harris A, Seckl J (2011) Glucocorticoids, prenatal stress and the programming of disease. Horm Behav 59: 279–289. - PubMed
-
- Veru F, Laplante DP, Luheshi G, King S (2014) Prenatal maternal stress exposure and immune function in the offspring. Stress 17: 133–148. - PubMed
-
- Coe CL, Lubach GR (2008) Fetal Programming: Prenatal Origins of Health and Illness. Current Directions in Psychological Science 17: 36–41.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

