Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea
- PMID: 25238399
- PMCID: PMC4303628
- DOI: 10.1038/ismej.2014.129
Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea
Abstract
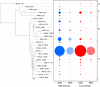
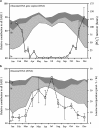
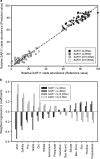
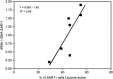
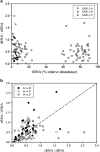
A seven-year oceanographic time series in NW Mediterranean surface waters was combined with pyrosequencing of ribosomal RNA (16S rRNA) and ribosomal RNA gene copies (16S rDNA) to examine the environmental controls on SAR11 ecotype dynamics and potential activity. SAR11 diversity exhibited pronounced seasonal cycles remarkably similar to total bacterial diversity. The timing of diversity maxima was similar across narrow and broad phylogenetic clades and strongly associated with deep winter mixing. Diversity minima were associated with periods of stratification that were low in nutrients and phytoplankton biomass and characterised by intense phosphate limitation (turnover time<5 h). We propose a conceptual framework in which physical mixing of the water column periodically resets SAR11 communities to a high diversity state and the seasonal evolution of phosphate limitation competitively excludes deeper-dwelling ecotypes to promote low diversity states dominated (>80%) by SAR11 Ia. A partial least squares (PLS) regression model was developed that could reliably predict sequence abundances of SAR11 ecotypes (Q(2)=0.70) from measured environmental variables, of which mixed layer depth was quantitatively the most important. Comparison of clade-level SAR11 rRNA:rDNA signals with leucine incorporation enabled us to partially validate the use of these ratios as an in-situ activity measure. However, temporal trends in the activity of SAR11 ecotypes and their relationship to environmental variables were unclear. The strong and predictable temporal patterns observed in SAR11 sequence abundance was not linked to metabolic activity of different ecotypes at the phylogenetic and temporal resolution of our study.
Figures

References
-
- Alonso-Sáez LV, Balagué E, Sà O, Sánchez JM, González J, Pinhassi R, et al. Seasonality in bacterial diversity in NW Mediterranean coastal waters: assessment through clone libraries, fingerprinting and fluorescence in situ hybridization. FEMS Microbiol Ecol. 2007;60:98–112. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
-
- Blanquer A, Uriz MJ, Galand PE. Removing environmental sources of variation to gain insight on symbionts vs. transient microbes in high and low microbial abundance sponges. Environ Microbiol. 2013;15:3008–3019. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

