Novel fusion transcripts associate with progressive prostate cancer
- PMID: 25238935
- PMCID: PMC4188871
- DOI: 10.1016/j.ajpath.2014.06.025
Novel fusion transcripts associate with progressive prostate cancer
Abstract
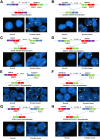
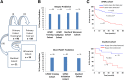
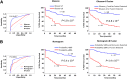
The mechanisms underlying the potential for aggressive behavior of prostate cancer (PCa) remain elusive. In this study, whole genome and/or transcriptome sequencing was performed on 19 specimens of PCa, matched adjacent benign prostate tissues, matched blood specimens, and organ donor prostates. A set of novel fusion transcripts was discovered in PCa. Eight of these fusion transcripts were validated through multiple approaches. The occurrence of these fusion transcripts was then analyzed in 289 prostate samples from three institutes, with clinical follow-up ranging from 1 to 15 years. The analyses indicated that most patients [69 (91%) of 76] positive for any of these fusion transcripts (TRMT11-GRIK2, SLC45A2-AMACR, MTOR-TP53BP1, LRRC59-FLJ60017, TMEM135-CCDC67, KDM4-AC011523.2, MAN2A1-FER, and CCNH-C5orf30) experienced PCa recurrence, metastases, and/or PCa-specific death after radical prostatectomy. These outcomes occurred in only 37% (58/157) of patients without carrying those fusion transcripts. Three fusion transcripts occurred exclusively in PCa samples from patients who experienced recurrence or PCaerelated death. The formation of these fusion transcripts may be the result of genome recombination. A combination of these fusion transcripts in PCa with Gleason's grading or with nomogram significantly improves the prediction rate of PCa recurrence. Our analyses suggest that formation of these fusion transcripts may underlie the aggressive behavior of PCa.
Figures


Comment in
-
Fusing transcriptomics to progressive prostate cancer.Am J Pathol. 2014 Oct;184(10):2608-10. doi: 10.1016/j.ajpath.2014.08.001. Epub 2014 Aug 14. Am J Pathol. 2014. PMID: 25128905
Similar articles
-
Discovery and Classification of Fusion Transcripts in Prostate Cancer and Normal Prostate Tissue.Am J Pathol. 2015 Jul;185(7):1834-45. doi: 10.1016/j.ajpath.2015.03.008. Epub 2015 May 9. Am J Pathol. 2015. PMID: 25963990 Free PMC article. Review.
-
Detection of fusion gene transcripts in the blood samples of prostate cancer patients.Sci Rep. 2021 Aug 20;11(1):16995. doi: 10.1038/s41598-021-96528-9. Sci Rep. 2021. PMID: 34417538 Free PMC article.
-
Identification of recurrent fusion genes across multiple cancer types.Sci Rep. 2019 Jan 31;9(1):1074. doi: 10.1038/s41598-019-38550-6. Sci Rep. 2019. PMID: 30705370 Free PMC article.
-
Characterization and evaluation of gene fusions as a measure of genetic instability and disease prognosis in prostate cancer.BMC Cancer. 2023 Jun 22;23(1):575. doi: 10.1186/s12885-023-11019-6. BMC Cancer. 2023. PMID: 37349736 Free PMC article.
-
The prognostic and predictive value of TMPRSS2-ERG gene fusion and ERG protein expression in prostate cancer biopsies.Dan Med J. 2016 Dec;63(12):B5319. Dan Med J. 2016. PMID: 27910803 Review.
Cited by
-
IFDlong: an isoform and fusion detector for accurate annotation and quantification of long-read RNA-seq data.bioRxiv [Preprint]. 2024 May 14:2024.05.11.593690. doi: 10.1101/2024.05.11.593690. bioRxiv. 2024. PMID: 38798496 Free PMC article. Preprint.
-
Roles of transmembrane protein 135 in mitochondrial and peroxisomal functions - implications for age-related retinal disease.Front Ophthalmol (Lausanne). 2024;4:1355379. doi: 10.3389/fopht.2024.1355379. Epub 2024 Jan 31. Front Ophthalmol (Lausanne). 2024. PMID: 38576540 Free PMC article.
-
MAN2A1-FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice.Gastroenterology. 2017 Oct;153(4):1120-1132.e15. doi: 10.1053/j.gastro.2016.12.036. Epub 2017 Feb 27. Gastroenterology. 2017. PMID: 28245430 Free PMC article.
-
Discovery and Classification of Fusion Transcripts in Prostate Cancer and Normal Prostate Tissue.Am J Pathol. 2015 Jul;185(7):1834-45. doi: 10.1016/j.ajpath.2015.03.008. Epub 2015 May 9. Am J Pathol. 2015. PMID: 25963990 Free PMC article. Review.
-
Fusion Gene Detection in Prostate Cancer Samples Enhances the Prediction of Prostate Cancer Clinical Outcomes from Radical Prostatectomy through Machine Learning in a Multi-Institutional Analysis.Am J Pathol. 2023 Apr;193(4):392-403. doi: 10.1016/j.ajpath.2022.12.013. Epub 2023 Jan 18. Am J Pathol. 2023. PMID: 36681188 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

