A transcriptional regulator Sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803
- PMID: 25239498
- PMCID: PMC4256502
- DOI: 10.1074/mcp.M113.035675
A transcriptional regulator Sll0794 regulates tolerance to biofuel ethanol in photosynthetic Synechocystis sp. PCC 6803
Abstract
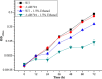
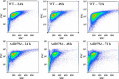
To improve ethanol production directly from CO2 in photosynthetic cyanobacterial systems, one key issue that needs to be addressed is the low ethanol tolerance of cyanobacterial cells. Our previous proteomic and transcriptomic analyses found that several regulatory proteins were up-regulated by exogenous ethanol in Synechocystis sp. PCC6803. In this study, through tolerance analysis of the gene disruption mutants of the up-regulated regulatory genes, we uncovered that one transcriptional regulator, Sll0794, was related directly to ethanol tolerance in Synechocystis. Using a quantitative iTRAQ-LC-MS/MS proteomics approach coupled with quantitative real-time reverse transcription-PCR (RT-qPCR), we further determined the possible regulatory network of Sll0794. The proteomic analysis showed that in the Δsll0794 mutant grown under ethanol stress a total of 54 and 87 unique proteins were down- and up-regulated, respectively. In addition, electrophoretic mobility shift assays demonstrated that the Sll0794 transcriptional regulator was able to bind directly to the upstream regions of sll1514, slr1512, and slr1838, which encode a 16.6 kDa small heat shock protein, a putative sodium-dependent bicarbonate transporter and a carbon dioxide concentrating mechanism protein CcmK, respectively. The study provided a proteomic description of the putative ethanol-tolerance network regulated by the sll0794 gene, and revealed new insights on the ethanol-tolerance regulatory mechanism in Synechocystis. As the first regulatory protein discovered related to ethanol tolerance, the gene may serve as a valuable target for transcription machinery engineering to further improve ethanol tolerance in Synechocystis. All MS data have been deposited in the ProteomeXchange with identifier PXD001266 (http://proteomecentral.proteomexchange.org/dataset/PXD001266).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
An orphan response regulator Sll0649 involved in cadmium tolerance and metal homeostasis in photosynthetic Synechocystis sp. PCC 6803.J Proteomics. 2014 May 30;103:87-102. doi: 10.1016/j.jprot.2014.03.029. Epub 2014 Apr 3. J Proteomics. 2014. PMID: 24704854
-
Integrated proteomic and metabolomic characterization of a novel two-component response regulator Slr1909 involved in acid tolerance in Synechocystis sp. PCC 6803.J Proteomics. 2014 Sep 23;109:76-89. doi: 10.1016/j.jprot.2014.06.021. Epub 2014 Jul 3. J Proteomics. 2014. PMID: 24998436
-
Integrated proteomic and transcriptomic analysis reveals novel genes and regulatory mechanisms involved in salt stress responses in Synechocystis sp. PCC 6803.Appl Microbiol Biotechnol. 2013 Sep;97(18):8253-64. doi: 10.1007/s00253-013-5139-8. Epub 2013 Aug 8. Appl Microbiol Biotechnol. 2013. PMID: 23925534
-
Quantitative iTRAQ LC-MS/MS proteomics reveals metabolic responses to biofuel ethanol in cyanobacterial Synechocystis sp. PCC 6803.J Proteome Res. 2012 Nov 2;11(11):5286-300. doi: 10.1021/pr300504w. Epub 2012 Oct 23. J Proteome Res. 2012. PMID: 23062023
-
Synechocystis: Not Just a Plug-Bug for CO2, but a Green E. coli.Front Bioeng Biotechnol. 2014 Sep 18;2:36. doi: 10.3389/fbioe.2014.00036. eCollection 2014. Front Bioeng Biotechnol. 2014. PMID: 25279375 Free PMC article. Review.
Cited by
-
Evaluation of the ethanol tolerance for wild and mutant Synechocystis strains by flow cytometry.Biotechnol Rep (Amst). 2018 Feb 15;17:137-147. doi: 10.1016/j.btre.2018.02.005. eCollection 2018 Mar. Biotechnol Rep (Amst). 2018. PMID: 29556479 Free PMC article.
-
Regulation Mechanism Mediated by Trans-Encoded sRNA Nc117 in Short Chain Alcohols Tolerance in Synechocystis sp. PCC 6803.Front Microbiol. 2018 May 1;9:863. doi: 10.3389/fmicb.2018.00863. eCollection 2018. Front Microbiol. 2018. PMID: 29780373 Free PMC article.
-
A novel small RNA CoaR regulates coenzyme A biosynthesis and tolerance of Synechocystis sp. PCC6803 to 1-butanol possibly via promoter-directed transcriptional silencing.Biotechnol Biofuels. 2017 Feb 20;10:42. doi: 10.1186/s13068-017-0727-y. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28239414 Free PMC article.
-
Expression of tardigrade disordered proteins impacts the tolerance to biofuels in a model cyanobacterium Synechocystis sp. PCC 6803.Front Microbiol. 2023 Jan 4;13:1091502. doi: 10.3389/fmicb.2022.1091502. eCollection 2022. Front Microbiol. 2023. PMID: 36687595 Free PMC article.
-
Bioethanol production from microalgae polysaccharides.Folia Microbiol (Praha). 2019 Sep;64(5):627-644. doi: 10.1007/s12223-019-00732-0. Epub 2019 Jul 27. Folia Microbiol (Praha). 2019. PMID: 31352666 Review.
References
-
- Antoni D., Zverlov V. V., Schwarz W. H. (2007) Biofuels from microbes. Appl. Microbiol. Biotechnol. 77, 23–35 - PubMed
-
- Renewable Fuels Association (2012) “Acelerating Industry Innovation – 2012 Ethanol Industry Outlook.” Renewable Fuels Association. pp. 3, 8,, 10,, 22,, and 23
-
- Shao X., Raman B., Zhu M., Mielenz J. R., Brown S. D., Guss A. M., Lynd L. R. (2011) Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 92, 641–652 - PubMed
-
- Kim H. S., Kim N. R., Yang J., Choi W. (2011) Identification of novel genes responsible for ethanol and/or thermotolerance by transposon mutagenesis in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 91, 1159–1172 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

