Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis
- PMID: 25239623
- PMCID: PMC4231657
- DOI: 10.1074/jbc.M114.573246
Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis
Abstract
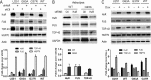
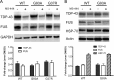
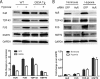
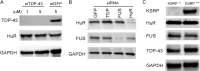
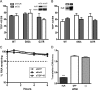
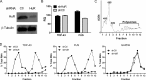
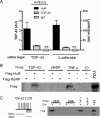
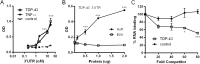
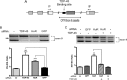
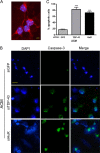
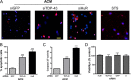
Posttranscriptional gene regulation is governed by a network of RNA-binding proteins (RBPs) that interact with regulatory elements in the mRNA to modulate multiple molecular processes, including splicing, RNA transport, RNA stability, and translation. Mounting evidence indicates that there is a hierarchy within this network whereby certain RBPs cross-regulate other RBPs to coordinate gene expression. HuR, an RNA-binding protein we linked previously to aberrant VEGF mRNA metabolism in models of SOD1-associated amyotrophic lateral sclerosis, has been identified as being high up in this hierarchy, serving as a regulator of RNA regulators. Here we investigated the role of HuR in regulating two RBPs, TDP-43 and FUS/TLS, that have been linked genetically to amyotrophic lateral sclerosis. We found that HuR promotes the expression of both RBPs in primary astrocytes and U251 cells under normal and stressed (hypoxic) conditions. For TDP-43, we found that HuR binds to the 3' untranslated region (UTR) and regulates its expression through translational efficiency rather than RNA stability. With HuR knockdown, there was a shift of TDP-43 and FUS mRNAs away from polysomes, consistent with translational silencing. The TDP-43 splicing function was attenuated upon HuR knockdown and could be rescued by ectopic TDP-43 lacking the 3' UTR regulatory elements. Finally, conditioned medium from astrocytes in which HuR or TDP-43 was knocked down produced significant motor neuron and cortical neuron toxicity in vitro. These findings indicate that HuR regulates TDP-43 and FUS/TLS expression and that loss of HuR-mediated RNA processing in astrocytes can alter the molecular and cellular landscape to produce a toxic phenotype.
Keywords: 3′ Untranslated Region; Amyotrophic Lateral Sclerosis (ALS) (Lou Gehrig Disease); Astrocyte; Neuron; RNA Processing; RNA Splicing.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs.Nat Neurosci. 2012 Nov;15(11):1488-97. doi: 10.1038/nn.3230. Epub 2012 Sep 30. Nat Neurosci. 2012. PMID: 23023293 Free PMC article.
-
Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA.J Biol Chem. 2010 Oct 29;285(44):34097-105. doi: 10.1074/jbc.M110.154831. Epub 2010 Aug 18. J Biol Chem. 2010. PMID: 20720006 Free PMC article.
-
TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells.J Biol Chem. 2012 May 4;287(19):15635-47. doi: 10.1074/jbc.M111.333450. Epub 2012 Mar 16. J Biol Chem. 2012. PMID: 22427648 Free PMC article.
-
Understanding the role of TDP-43 and FUS/TLS in ALS and beyond.Curr Opin Neurobiol. 2011 Dec;21(6):904-19. doi: 10.1016/j.conb.2011.05.029. Epub 2011 Aug 2. Curr Opin Neurobiol. 2011. PMID: 21813273 Free PMC article. Review.
-
Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins.J Neurochem. 2016 Aug;138 Suppl 1:95-111. doi: 10.1111/jnc.13625. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27015757 Review.
Cited by
-
RNA-Targeted Therapies and High-Throughput Screening Methods.Int J Mol Sci. 2020 Apr 23;21(8):2996. doi: 10.3390/ijms21082996. Int J Mol Sci. 2020. PMID: 32340368 Free PMC article. Review.
-
Neuron-Specific HuR-Deficient Mice Spontaneously Develop Motor Neuron Disease.J Immunol. 2018 Jul 1;201(1):157-166. doi: 10.4049/jimmunol.1701501. Epub 2018 May 14. J Immunol. 2018. PMID: 29760195 Free PMC article.
-
HuR-Dependent Editing of a New Mineralocorticoid Receptor Splice Variant Reveals an Osmoregulatory Loop for Sodium Homeostasis.Sci Rep. 2017 Jul 6;7(1):4835. doi: 10.1038/s41598-017-04838-8. Sci Rep. 2017. PMID: 28684740 Free PMC article.
-
A Requirement for Mena, an Actin Regulator, in Local mRNA Translation in Developing Neurons.Neuron. 2017 Aug 2;95(3):608-622.e5. doi: 10.1016/j.neuron.2017.06.048. Epub 2017 Jul 20. Neuron. 2017. PMID: 28735747 Free PMC article.
-
Transactive response DNA-binding protein 43 is enriched at the centrosome in human cells.Brain. 2023 Sep 1;146(9):3624-3633. doi: 10.1093/brain/awad228. Brain. 2023. PMID: 37410912 Free PMC article.
References
-
- Robberecht W., Philips T. (2013) The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 14, 248–264 - PubMed
-
- Buratti E., Baralle F. E. (2011) TDP-43: new aspects of autoregulation mechanisms in RNA binding proteins and their connection with human disease. FEBS J. 278, 3530–3538 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

