Generation of the epicardial lineage from human pluripotent stem cells
- PMID: 25240927
- PMCID: PMC4192149
- DOI: 10.1038/nbt.3002
Generation of the epicardial lineage from human pluripotent stem cells
Abstract
The epicardium supports cardiomyocyte proliferation early in development and provides fibroblasts and vascular smooth muscle cells to the developing heart. The epicardium has been shown to play an important role during tissue remodeling after cardiac injury, making access to this cell lineage necessary for the study of regenerative medicine. Here we describe the generation of epicardial lineage cells from human pluripotent stem cells by stage-specific activation of the BMP and WNT signaling pathways. These cells display morphological characteristics and express markers of the epicardial lineage, including the transcription factors WT1 and TBX18 and the retinoic acid-producing enzyme ALDH1A2. When induced to undergo epithelial-to-mesenchymal transition, the cells give rise to populations that display characteristics of the fibroblast and vascular smooth muscle lineages. These findings identify BMP and WNT as key regulators of the epicardial lineage in vitro and provide a model for investigating epicardial function in human development and disease.
Figures

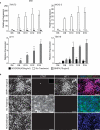
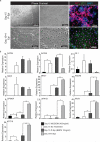
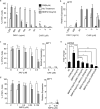
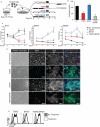
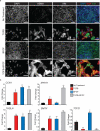
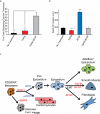
Comment in
-
Pluripotent-stem-cell-derived epicardial cells: a step toward artificial cardiac tissue.Cell Stem Cell. 2014 Nov 6;15(5):533-4. doi: 10.1016/j.stem.2014.10.007. Epub 2014 Nov 6. Cell Stem Cell. 2014. PMID: 25517459
References
-
- DeRuiter MC, Poelmann RE, VanderPlas-de Vries I, Mentink MM, Gittenberger-de Groot AC. The development of the myocardium and endocardium in mouse embryos. Fusion of two heart tubes? Anatomy and embryology. 1992;185:461–473. - PubMed
-
- Limana F, Capogrossi MC, Germani A. The epicardium in cardiac repair: from the stem cell view. Pharmacology & therapeutics. 2011;129:82–96. - PubMed
-
- Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anatomy and embryology. 1987;176:183–189. - PubMed
-
- Austin AF, Compton LA, Love JD, Brown CB, Barnett JV. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev Dyn. 2008;237:366–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

