Epigenetic inheritance: histone bookmarks across generations
- PMID: 25242115
- PMCID: PMC4254315
- DOI: 10.1016/j.tcb.2014.08.004
Epigenetic inheritance: histone bookmarks across generations
Abstract
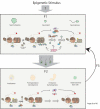
Multiple circuitries ensure that cells respond correctly to the environmental cues within defined cellular programs. There is increasing evidence suggesting that cellular memory for these adaptive processes can be passed on through cell divisions and generations. However, the mechanisms by which this epigenetic information is transferred remain elusive, largely because it requires that such memory survive through gross chromatin remodeling events during DNA replication, mitosis, meiosis, and developmental reprogramming. Elucidating the processes by which epigenetic information survives and is transmitted is a central challenge in biology. In this review, we consider recent advances in understanding mechanisms of epigenetic inheritance with a focus on histone segregation at the replication fork, and how an epigenetic memory may get passed through the paternal lineage.
Keywords: epigenetic; gamete; histone; inheritance; polycomb; replication.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
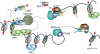
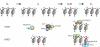
References
-
- Annunziato AT. Assembling chromatin: The long and winding road. Biochim Biophys Acta. 2012;1819:196–210. - PubMed
-
- Herman TM, et al. Structure of chromatin at deoxyribonucleic acid replication forks: location of the first nucleosomes on newly synthesized simian virus 40 deoxyribonucleic acid. Biochemistry. 1981;20:621–630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

