Novel DNA methylation targets in oral rinse samples predict survival of patients with oral squamous cell carcinoma
- PMID: 25242135
- PMCID: PMC4254027
- DOI: 10.1016/j.oraloncology.2014.08.015
Novel DNA methylation targets in oral rinse samples predict survival of patients with oral squamous cell carcinoma
Abstract
Objectives: The objective of this study was to identify novel survival-associated biomarkers in oral rinse samples collected from patients with oral squamous cell carcinoma (OSCC).
Materials and methods: We screened for putative survival-associated markers using publicly available methylation array data from 88 OSCC tumors. Cox models were then fit to methylation array data restricted to these putative loci in oral rinse samples of 82 OSCC patients from greater Boston. Pyrosequencing assays were designed for each locus that replicated in the oral rinse samples and applied to a validation set of oral rinse samples from another 61 OSCC patients.
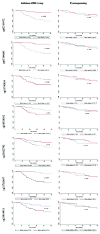
Results: We identified 7 survival-associated methylation markers in oral rinse samples from OSCC patients, and have validated one, located in the body of GABBR1, by pyrosequencing.
Conclusion: The 7 CpG loci identified through this study represent novel prognostic biomarkers for patients with OSCC that can be detected using a non-invasive oral rinse collection technique.
Keywords: Epigenomics; GABBR1; Mouthwash; Oral cancer; Prognosis.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Conflict of interest statement
We have no conflicts to declare.
Figures


Similar articles
-
TRH site-specific methylation in oral and oropharyngeal squamous cell carcinoma.BMC Cancer. 2018 Aug 6;18(1):786. doi: 10.1186/s12885-018-4706-x. BMC Cancer. 2018. PMID: 30081853 Free PMC article.
-
Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India.Clin Epigenetics. 2017 Feb 3;9:13. doi: 10.1186/s13148-017-0314-x. eCollection 2017. Clin Epigenetics. 2017. PMID: 28174608 Free PMC article.
-
An integrated methylation and gene expression microarray analysis reveals significant prognostic biomarkers in oral squamous cell carcinoma.Oncol Rep. 2018 Nov;40(5):2637-2647. doi: 10.3892/or.2018.6702. Epub 2018 Sep 12. Oncol Rep. 2018. PMID: 30226546 Free PMC article.
-
Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma.Clin Epigenetics. 2017 Aug 24;9:88. doi: 10.1186/s13148-017-0392-9. eCollection 2017. Clin Epigenetics. 2017. PMID: 28852427 Free PMC article.
-
Aberrant DNA methylation of tumor-related genes in oral rinse: a noninvasive method for detection of oral squamous cell carcinoma.Cancer. 2012 Sep 1;118(17):4298-308. doi: 10.1002/cncr.27417. Epub 2012 Jan 17. Cancer. 2012. PMID: 22252571
Cited by
-
High expression of vinculin predicts poor prognosis and distant metastasis and associates with influencing tumor-associated NK cell infiltration and epithelial-mesenchymal transition in gastric cancer.Aging (Albany NY). 2021 Feb 1;13(4):5197-5225. doi: 10.18632/aging.202440. Epub 2021 Feb 1. Aging (Albany NY). 2021. PMID: 33535187 Free PMC article.
-
ATHENA: an independently validated autophagy-related epigenetic prognostic prediction model of head and neck squamous cell carcinoma.Clin Epigenetics. 2023 Jun 9;15(1):97. doi: 10.1186/s13148-023-01501-0. Clin Epigenetics. 2023. PMID: 37296474 Free PMC article.
-
Saliva Gene Promoter Hypermethylation as a Biomarker in Oral Cancer.J Clin Med. 2021 Apr 29;10(9):1931. doi: 10.3390/jcm10091931. J Clin Med. 2021. PMID: 33947071 Free PMC article. Review.
-
Unidirectional alteration of methylation and hydroxymethylation at the promoters and differential gene expression in oral squamous cell carcinoma.Front Genet. 2023 Oct 12;14:1269084. doi: 10.3389/fgene.2023.1269084. eCollection 2023. Front Genet. 2023. PMID: 37900177 Free PMC article.
-
Differences in the prognosis of gastric cancer patients of different sexes and races and the molecular mechanisms involved.Int J Oncol. 2019 Nov;55(5):1049-1068. doi: 10.3892/ijo.2019.4885. Epub 2019 Sep 26. Int J Oncol. 2019. PMID: 31793655 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. - PubMed
-
- Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114:806–16. - PubMed
-
- Altekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review, 1975-2007. Bethesda, MD: National Cancer Institute; 2010.
-
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

