Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion
- PMID: 25242145
- PMCID: PMC4224619
- DOI: 10.1016/j.molcel.2014.08.018
Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion
Abstract
Many cancer cells consume large quantities of glutamine to maintain TCA cycle anaplerosis and support cell survival. It was therefore surprising when RNAi screening revealed that suppression of citrate synthase (CS), the first TCA cycle enzyme, prevented glutamine-withdrawal-induced apoptosis. CS suppression reduced TCA cycle activity and diverted oxaloacetate, the substrate of CS, into production of the nonessential amino acids aspartate and asparagine. We found that asparagine was necessary and sufficient to suppress glutamine-withdrawal-induced apoptosis without restoring the levels of other nonessential amino acids or TCA cycle intermediates. In complete medium, tumor cells exhibiting high rates of glutamine consumption underwent rapid apoptosis when glutamine-dependent asparagine synthesis was suppressed, and expression of asparagine synthetase was statistically correlated with poor prognosis in human tumors. Coupled with the success of L-asparaginase as a therapy for childhood leukemia, the data suggest that intracellular asparagine is a critical suppressor of apoptosis in many human tumors.
Figures
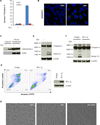

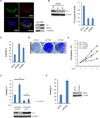
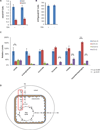

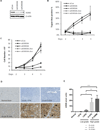
References
-
- Avramis VI. Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res. 2012;32:2423–2437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

