Growing knowledge of the mTOR signaling network
- PMID: 25242279
- PMCID: PMC4253687
- DOI: 10.1016/j.semcdb.2014.09.011
Growing knowledge of the mTOR signaling network
Abstract
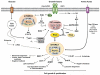
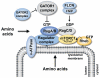
The kinase mTOR (mechanistic target of rapamycin) integrates diverse environmental signals and translates these cues into appropriate cellular responses. mTOR forms the catalytic core of at least two functionally distinct signaling complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 promotes anabolic cellular metabolism in response to growth factors, nutrients, and energy and functions as a master controller of cell growth. While significantly less well understood than mTORC1, mTORC2 responds to growth factors and controls cell metabolism, cell survival, and the organization of the actin cytoskeleton. mTOR plays critical roles in cellular processes related to tumorigenesis, metabolism, immune function, and aging. Consequently, aberrant mTOR signaling contributes to myriad disease states, and physicians employ mTORC1 inhibitors (rapamycin and analogs) for several pathological conditions. The clinical utility of mTOR inhibition underscores the important role of mTOR in organismal physiology. Here we review our growing knowledge of cellular mTOR regulation by diverse upstream signals (e.g. growth factors; amino acids; energy) and how mTORC1 integrates these signals to effect appropriate downstream signaling, with a greater emphasis on mTORC1 over mTORC2. We highlight dynamic subcellular localization of mTORC1 and associated factors as an important mechanism for control of mTORC1 activity and function. We will cover major cellular functions controlled by mTORC1 broadly. While significant advances have been made in the last decade regarding the regulation and function of mTOR within complex cell signaling networks, many important findings remain to be discovered.
Keywords: Amino acids; Energy; Insulin; mTOR; mTORC1; mTORC2.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

References
-
- Abraham RT, Wiederrecht GJ. Immunopharmacology of rapamycin. Annu Rev Immunol. 1996;14:483–510. PMID. - PubMed
-
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. PMID: 15094765. - PubMed
-
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. PMID: 1715094. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

