Brucella papionis sp. nov., isolated from baboons (Papio spp.)
- PMID: 25242540
- PMCID: PMC4811636
- DOI: 10.1099/ijs.0.065482-0
Brucella papionis sp. nov., isolated from baboons (Papio spp.)
Abstract
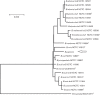
Two Gram-negative, non-motile, non-spore-forming coccoid bacteria (strains F8/08-60(T) and F8/08-61) isolated from clinical specimens obtained from baboons (Papio spp.) that had delivered stillborn offspring were subjected to a polyphasic taxonomic study. On the basis of 16S rRNA gene sequence similarities, both strains, which possessed identical sequences, were assigned to the genus Brucella. This placement was confirmed by extended multilocus sequence analysis (MLSA), where both strains possessed identical sequences, and whole-genome sequencing of a representative isolate. All of the above analyses suggested that the two strains represent a novel lineage within the genus Brucella. The strains also possessed a unique profile when subjected to the phenotyping approach classically used to separate species of the genus Brucella, reacting only with Brucella A monospecific antiserum, being sensitive to the dyes thionin and fuchsin, being lysed by bacteriophage Wb, Bk2 and Fi phage at routine test dilution (RTD) but only partially sensitive to bacteriophage Tb, and with no requirement for CO2 and no production of H2S but strong urease activity. Biochemical profiling revealed a pattern of enzyme activity and metabolic capabilities distinct from existing species of the genus Brucella. Molecular analysis of the omp2 locus genes showed that both strains had a novel combination of two highly similar omp2b gene copies. The two strains shared a unique fingerprint profile of the multiple-copy Brucella-specific element IS711. Like MLSA, a multilocus variable number of tandem repeat analysis (MLVA) showed that the isolates clustered together very closely, but represent a distinct group within the genus Brucella. Isolates F8/08-60(T) and F8/08-61 could be distinguished clearly from all known species of the genus Brucella and their biovars by both phenotypic and molecular properties. Therefore, by applying the species concept for the genus Brucella suggested by the ICSP Subcommittee on the Taxonomy of Brucella, they represent a novel species within the genus Brucella, for which the name Brucella papionis sp. nov. is proposed, with the type strain F8/08-60(T) ( = NCTC 13660(T) = CIRMBP 0958(T)).
Crown Copyright 2014. Reproduced with the permission of the Controller of Her Majesty's Stationery Office/Queen's Printer for Scotland and AHVLA.
Figures

References
-
- Al Dahouk S., Hofer E., Tomaso H., Vergnaud G., Le Flèche P., Cloeckaert A., Koylass M. S., Whatmore A. M., Nöckler K., Scholz H. C. (2012). Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl Environ Microbiol 78, 1534–1543. 10.1128/AEM.06351-11 - DOI - PMC - PubMed
-
- Alton G. G., Jones L. M., Angus R. D., Verger J. M. (1988). Techniques for the Brucellosis Laboratory. Paris: INRA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

