High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis
- PMID: 25244001
- PMCID: PMC4245175
- DOI: 10.1021/cb500622c
High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis
Abstract
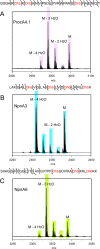
Lanthionine-containing peptides (lanthipeptides) are a rapidly growing family of polycyclic peptide natural products belonging to the large class of ribosomally synthesized and post-translationally modified peptides (RiPPs). These compounds are widely distributed in taxonomically distant species, and their biosynthetic systems and biological activities are diverse. A unique example of lanthipeptide biosynthesis is the prochlorosin synthetase ProcM from the marine cyanobacterium Prochlorococcus MIT9313, which transforms up to 29 different precursor peptides (ProcAs) into a library of lanthipeptides called prochlorosins (Pcns) with highly diverse sequences and ring topologies. Here, we show that many ProcM-like enzymes from a variety of bacteria have the capacity to carry out post-translational modifications on highly diverse precursor peptides, providing new examples of natural combinatorial biosynthesis. We also demonstrate that the leader peptides come from different evolutionary origins, suggesting that the combinatorial biosynthesis is tied to the enzyme and not a specific type of leader peptide. For some precursor peptides encoded in the genomes, the leader peptides apparently have been truncated at the N-termini, and we show that these N-terminally truncated peptides are still substrates of the enzymes. Consistent with this hypothesis, we demonstrate that about two-thirds of the ProcA N-terminal sequence is not essential for ProcM activity. Our results also highlight the potential of exploring this class of natural products by genome mining and bioengineering.
Figures





References
-
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.-D.; Fischbach M. A.; Garavelli J. S.; Göransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Müller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J. T.; Rebuffat S.; Ross R. P.; Sahl H.-G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Süssmuth R. E.; Tagg J. R.; Tang G.-L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. - PMC - PubMed
-
- Willey J. M.; van der Donk W. A. (2007) Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501. - PubMed
-
- Knerr P. J.; van der Donk W. A. (2012) Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505. - PubMed
-
- Bierbaum G.; Sahl H. G. (2009) Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10, 2–18. - PubMed
-
- Piper C.; Cotter P. D.; Ross R. P.; Hill C. (2009) Discovery of medically significant lantibiotics. Curr. Drug Discovery Technol. 6, 1–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

