Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity
- PMID: 25245761
- PMCID: PMC4203949
- DOI: 10.1084/jem.20141308
Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity
Abstract
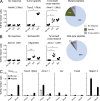
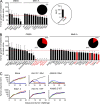
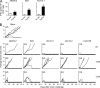
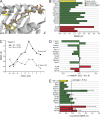
The mutational repertoire of cancers creates the neoepitopes that make cancers immunogenic. Here, we introduce two novel tools that identify, with relatively high accuracy, the small proportion of neoepitopes (among the hundreds of potential neoepitopes) that protect the host through an antitumor T cell response. The two tools consist of (a) the numerical difference in NetMHC scores between the mutated sequences and their unmutated counterparts, termed the differential agretopic index, and (b) the conformational stability of the MHC I-peptide interaction. Mechanistically, these tools identify neoepitopes that are mutated to create new anchor residues for MHC binding, and render the overall peptide more rigid. Surprisingly, the protective neoepitopes identified here elicit CD8-dependent immunity, even though their affinity for K(d) is orders of magnitude lower than the 500-nM threshold considered reasonable for such interactions. These results greatly expand the universe of target cancer antigens and identify new tools for human cancer immunotherapy.
© 2014 Duan et al.
Figures
References
-
- Assarsson, E., Sidney J., Oseroff C., Pasquetto V., Bui H.H., Frahm N., Brander C., Peters B., Grey H., and Sette A.. 2007. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 178:7890–7901. 10.4049/jimmunol.178.12.7890 - DOI - PubMed
-
- Bailey, D.W.1978. Sources of subline divergences and their relative importance for sublines of six major inbred strains of mice. Origins of inbred mice. Morse H.C. III, editor. Elsevier; 10.1016/B978-0-12-507850-4.50020-2 - DOI
-
- Borbulevych, O.Y., Baxter T.K., Yu Z., Restifo N.P., and Baker B.M.. 2005. Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J. Immunol. 174:4812–4820. 10.4049/jimmunol.174.8.4812 - DOI - PMC - PubMed
-
- Borbulevych, O.Y., Insaidoo F.K., Baxter T.K., Powell D.J. Jr, Johnson L.A., Restifo N.P., and Baker B.M.. 2007. Structures of MART-126/27-35 Peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J. Mol. Biol. 372:1123–1136. 10.1016/j.jmb.2007.07.025 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

