A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction
- PMID: 25246656
- PMCID: PMC4201826
- DOI: 10.1261/rna.044362.114
A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction
Abstract
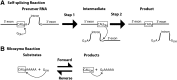
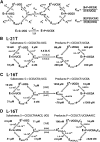
Determination of quantitative thermodynamic and kinetic frameworks for ribozymes derived from the Azoarcus group I intron and comparisons to their well-studied analogs from the Tetrahymena group I intron reveal similarities and differences between these RNAs. The guanosine (G) substrate binds to the Azoarcus and Tetrahymena ribozymes with similar equilibrium binding constants and similar very slow association rate constants. These and additional literature observations support a model in which the free ribozyme is not conformationally competent to bind G and in which the probability of assuming the binding-competent state is determined by tertiary interactions of peripheral elements. As proposed previously, the slow binding of guanosine may play a role in the specificity of group I intron self-splicing, and slow binding may be used analogously in other biological processes. The internal equilibrium between ribozyme-bound substrates and products is similar for these ribozymes, but the Azoarcus ribozyme does not display the coupling in the binding of substrates that is observed with the Tetrahymena ribozyme, suggesting that local preorganization of the active site and rearrangements within the active site upon substrate binding are different for these ribozymes. Our results also confirm the much greater tertiary binding energy of the 5'-splice site analog with the Azoarcus ribozyme, binding energy that presumably compensates for the fewer base-pairing interactions to allow the 5'-exon intermediate in self splicing to remain bound subsequent to 5'-exon cleavage and prior to exon ligation. Most generally, these frameworks provide a foundation for design and interpretation of experiments investigating fundamental properties of these and other structured RNAs.
Keywords: Group I intron; catalytic framework; noncoding RNA; ribozyme.
© 2014 Gleitsman and Herschlag; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



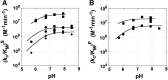
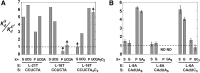
 , where KdX describes binding to the free ribozyme, while
, where KdX describes binding to the free ribozyme, while  refers to the binding in the presence of saturating concentrations of the other substrate/product. The black dashed line represents no coupling. The arrows signify values that are lower limits.
refers to the binding in the presence of saturating concentrations of the other substrate/product. The black dashed line represents no coupling. The arrows signify values that are lower limits.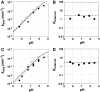
Similar articles
-
Trans-splicing with the group I intron ribozyme from Azoarcus.RNA. 2014 Feb;20(2):202-13. doi: 10.1261/rna.041012.113. Epub 2013 Dec 16. RNA. 2014. PMID: 24344321 Free PMC article.
-
A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme.J Mol Biol. 2000 Sep 15;302(2):339-58. doi: 10.1006/jmbi.2000.4056. J Mol Biol. 2000. PMID: 10970738
-
Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme.Biochim Biophys Acta. 1999 Dec 23;1489(2-3):281-92. doi: 10.1016/s0167-4781(99)00200-6. Biochim Biophys Acta. 1999. PMID: 10673029
-
Group I Intron-Based Therapeutics Through Trans-Splicing Reaction.Prog Mol Biol Transl Sci. 2018;159:79-100. doi: 10.1016/bs.pmbts.2018.07.001. Epub 2018 Aug 9. Prog Mol Biol Transl Sci. 2018. PMID: 30340790 Review.
-
In Vitro Self-Circularization Methods Based on Self-Splicing Ribozyme.Int J Mol Sci. 2024 Aug 30;25(17):9437. doi: 10.3390/ijms25179437. Int J Mol Sci. 2024. PMID: 39273386 Free PMC article. Review.
Cited by
-
Slow molecular recognition by RNA.RNA. 2017 Dec;23(12):1745-1753. doi: 10.1261/rna.062026.117. Epub 2017 Sep 28. RNA. 2017. PMID: 28971853 Free PMC article.
-
Dynamic RNA Fitness Landscapes of a Group I Ribozyme during Changes to the Experimental Environment.Mol Biol Evol. 2022 Mar 2;39(3):msab373. doi: 10.1093/molbev/msab373. Mol Biol Evol. 2022. PMID: 35020916 Free PMC article.
-
Mimicking Ribosomal Unfolding of RNA Pseudoknot in a Protein Channel.J Am Chem Soc. 2015 Dec 23;137(50):15742-52. doi: 10.1021/jacs.5b07910. Epub 2015 Dec 10. J Am Chem Soc. 2015. PMID: 26595106 Free PMC article.
-
Nucleobases Undergo Dynamic Rearrangements during RNA Tertiary Folding.J Mol Biol. 2016 Nov 6;428(22):4490-4502. doi: 10.1016/j.jmb.2016.09.015. Epub 2016 Sep 29. J Mol Biol. 2016. PMID: 27693721 Free PMC article.
-
Group I Intron Internal Guide Sequence Binding Strength as a Component of Ribozyme Network Formation.Molecules. 2016 Sep 27;21(10):1293. doi: 10.3390/molecules21101293. Molecules. 2016. PMID: 27689977 Free PMC article.
References
-
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. 2004b. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 - PubMed
-
- Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. 2003. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol 328: 1011–1026 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
