Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals
- PMID: 25247702
- PMCID: PMC4359377
- DOI: 10.7554/eLife.03640
Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals
Abstract
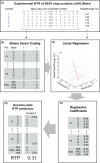
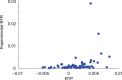
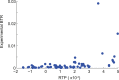
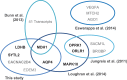
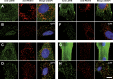
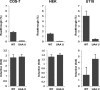
Translational readthrough gives rise to low abundance proteins with C-terminal extensions beyond the stop codon. To identify functional translational readthrough, we estimated the readthrough propensity (RTP) of all stop codon contexts of the human genome by a new regression model in silico, identified a nucleotide consensus motif for high RTP by using this model, and analyzed all readthrough extensions in silico with a new predictor for peroxisomal targeting signal type 1 (PTS1). Lactate dehydrogenase B (LDHB) showed the highest combined RTP and PTS1 probability. Experimentally we show that at least 1.6% of the total cellular LDHB is targeted to the peroxisome by a conserved hidden PTS1. The readthrough-extended lactate dehydrogenase subunit LDHBx can also co-import LDHA, the other LDH subunit, into peroxisomes. Peroxisomal LDH is conserved in mammals and likely contributes to redox equivalent regeneration in peroxisomes.
Keywords: cell biology; gene regulation; genome-wide in silico screen; human; human biology; lactate dehydrogenase; medicine; peroxisomal targeting signal; stop codon suppression; translational readthrough.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





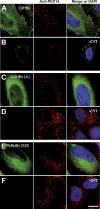
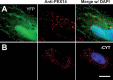
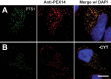
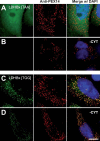




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

