Assessing the metabolic impact of nitrogen availability using a compartmentalized maize leaf genome-scale model
- PMID: 25248718
- PMCID: PMC4226342
- DOI: 10.1104/pp.114.245787
Assessing the metabolic impact of nitrogen availability using a compartmentalized maize leaf genome-scale model
Abstract
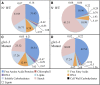
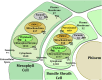
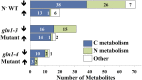
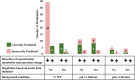
Maize (Zea mays) is an important C4 plant due to its widespread use as a cereal and energy crop. A second-generation genome-scale metabolic model for the maize leaf was created to capture C4 carbon fixation and investigate nitrogen (N) assimilation by modeling the interactions between the bundle sheath and mesophyll cells. The model contains gene-protein-reaction relationships, elemental and charge-balanced reactions, and incorporates experimental evidence pertaining to the biomass composition, compartmentalization, and flux constraints. Condition-specific biomass descriptions were introduced that account for amino acids, fatty acids, soluble sugars, proteins, chlorophyll, lignocellulose, and nucleic acids as experimentally measured biomass constituents. Compartmentalization of the model is based on proteomic/transcriptomic data and literature evidence. With the incorporation of information from the MetaCrop and MaizeCyc databases, this updated model spans 5,824 genes, 8,525 reactions, and 9,153 metabolites, an increase of approximately 4 times the size of the earlier iRS1563 model. Transcriptomic and proteomic data have also been used to introduce regulatory constraints in the model to simulate an N-limited condition and mutants deficient in glutamine synthetase, gln1-3 and gln1-4. Model-predicted results achieved 90% accuracy when comparing the wild type grown under an N-complete condition with the wild type grown under an N-deficient condition.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures
References
-
- Amiour N, Imbaud S, Clément G, Agier N, Zivy M, Valot B, Balliau T, Armengaud P, Quilleré I, Cañas R, et al. (2012) The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J Exp Bot 63: 5017–5033 - PubMed
-
- Amiour N, Imbaud S, Clément G, Agier N, Zivy M, Valot B, Balliau B, Quilleré I, Tercé-Laforgue T, Dargel-Graffin C, et al. (2014) An integrated “omics” approach to the characterization of maize (Zea mays L.) mutants deficient in the expression of two genes encoding cytosolic glutamine synthetase. BMC Genomics (in press) - PMC - PubMed
-
- Andrews M, Lea PJ. (2013) Our nitrogen ‘footprint’: the need for increased crop nitrogen use efficiency. Ann Appl Biol 163: 165–169
-
- Andrews M, Raven JA, Lea PJ. (2013) Do plants need nitrate? The mechanisms by which nitrogen form affects plants. Ann Appl Biol 163: 174–199
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

