Retinal mitochondrial DNA mismatch repair in the development of diabetic retinopathy, and its continued progression after termination of hyperglycemia
- PMID: 25249609
- PMCID: PMC4215746
- DOI: 10.1167/iovs.14-15020
Retinal mitochondrial DNA mismatch repair in the development of diabetic retinopathy, and its continued progression after termination of hyperglycemia
Abstract
Purpose: Mitochondrial DNA (mtDNA) is damaged in the retina in diabetes, and mitochondria copy numbers are decreased. The displacement-loop (D-loop) of the mtDNA, the region with transcription/replication elements, experiences more damage than other regions of mtDNA. Our aim was to examine the role of DNA mismatch repair (MMR) in mitochondria homeostasis in diabetic retinopathy, and in its continued progression after cessation of hyperglycemia.
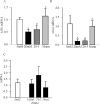
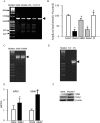
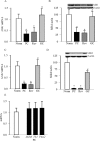
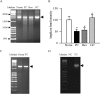
Methods: Effect of hyperglycemia on sequence variants in the D-loop region was investigated in retinal endothelial cells and in the retina from streptozotocin-induced diabetic rats using mismatch-specific surveyor nuclease. The role of MMR machinery in mtDNA damage and mitochondrial respiration was investigated in retinal endothelial cells overexpressing Mlh1, an MMR enzyme mainly associated with mtDNA polymerase gamma, or Msh2 (associated with nuclear polymerase beta).
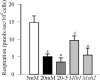
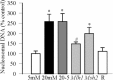
Results: Hyperglycemia increased sequence variants in the D-loop region. While overexpression of Mlh1 in endothelial cells ameliorated glucose-induced increase in D-loop sequence variants, decrease in respiration rate and increase in apoptosis, overexpression of Msh2 did not protect the mitochondria damage. Termination of hyperglycemia failed to reverse decrease in MMR enzymes and increase in D-loop sequence variants.
Conclusions: Due to a compromised MMR system, the sequence variants in the D-loop region were not repaired, and that resulted in impaired mtDNA transcription. Mitochondria become dysfunctional, and they continued to be dysfunctional even after hyperglycemia was terminated, contributing to the development, and progression of diabetic retinopathy. Thus, strategies targeting mitochondrial MMR machinery could help maintain mitochondria homeostasis, and inhibit the development of diabetic retinopathy and its continued progression.
Keywords: DNA repair; diabetic retinopathy; metabolic memory; mitochondria damage; mtDNA mismatch.
Copyright 2014 The Association for Research in Vision and Ophthalmology, Inc.
Figures
References
-
- Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci. 2003; 44: 5327–5334 - PubMed
-
- Kanwar M, Chan PS, Kern TS, Kowluru RA. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007; 48: 3805–3811 - PubMed
-
- Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005; 54: 1615–1625 - PubMed
-
- Stuart JA, Brown MF. Mitochondrial DNA maintenance and bioenergetics. Biochim Biophys Acta. 2006; 1757: 79–89 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

