Nova1 is a master regulator of alternative splicing in pancreatic beta cells
- PMID: 25249621
- PMCID: PMC4191425
- DOI: 10.1093/nar/gku861
Nova1 is a master regulator of alternative splicing in pancreatic beta cells
Abstract
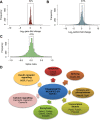
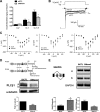
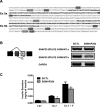
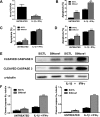
Alternative splicing (AS) is a fundamental mechanism for the regulation of gene expression. It affects more than 90% of human genes but its role in the regulation of pancreatic beta cells, the producers of insulin, remains unknown. Our recently published data indicated that the 'neuron-specific' Nova1 splicing factor is expressed in pancreatic beta cells. We have presently coupled specific knockdown (KD) of Nova1 with RNA-sequencing to determine all splice variants and downstream pathways regulated by this protein in beta cells. Nova1 KD altered the splicing of nearly 5000 transcripts. Pathway analysis indicated that these genes are involved in exocytosis, apoptosis, insulin receptor signaling, splicing and transcription. In line with these findings, Nova1 silencing inhibited insulin secretion and induced apoptosis basally and after cytokine treatment in rodent and human beta cells. These observations identify a novel layer of regulation of beta cell function, namely AS controlled by key splicing regulators such as Nova1.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Nova1 or Bim Deficiency in Pancreatic β-Cells Does Not Alter Multiple Low-Dose Streptozotocin-Induced Diabetes and Diet-Induced Obesity in Mice.Nutrients. 2022 Sep 18;14(18):3866. doi: 10.3390/nu14183866. Nutrients. 2022. PMID: 36145242 Free PMC article.
-
The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines.PLoS Genet. 2012;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412385 Free PMC article.
-
Neuron-enriched RNA-binding Proteins Regulate Pancreatic Beta Cell Function and Survival.J Biol Chem. 2017 Feb 24;292(8):3466-3480. doi: 10.1074/jbc.M116.748335. Epub 2017 Jan 11. J Biol Chem. 2017. PMID: 28077579 Free PMC article.
-
Alternative Splicing by NOVA Factors: From Gene Expression to Cell Physiology and Pathology.Int J Mol Sci. 2020 May 30;21(11):3941. doi: 10.3390/ijms21113941. Int J Mol Sci. 2020. PMID: 32486302 Free PMC article. Review.
-
Neuro-oncological ventral antigen 1 (NOVA1): Implications in neurological diseases and cancers.Cell Prolif. 2017 Aug;50(4):e12348. doi: 10.1111/cpr.12348. Epub 2017 Apr 10. Cell Prolif. 2017. PMID: 28394091 Free PMC article. Review.
Cited by
-
Stress-Induced Translational Regulation Mediated by RNA Binding Proteins: Key Links to β-Cell Failure in Diabetes.Diabetes. 2020 Apr;69(4):499-507. doi: 10.2337/dbi18-0068. Diabetes. 2020. PMID: 32198193 Free PMC article. Review.
-
Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α and β cells.Elife. 2015 Jun 10;4:e06990. doi: 10.7554/eLife.06990. Elife. 2015. PMID: 26061776 Free PMC article.
-
Thrombospondin 1 protects pancreatic β-cells from lipotoxicity via the PERK-NRF2 pathway.Cell Death Differ. 2016 Dec;23(12):1995-2006. doi: 10.1038/cdd.2016.89. Epub 2016 Sep 2. Cell Death Differ. 2016. PMID: 27588705 Free PMC article.
-
Interferons are key cytokines acting on pancreatic islets in type 1 diabetes.Diabetologia. 2024 May;67(5):908-927. doi: 10.1007/s00125-024-06106-7. Epub 2024 Feb 26. Diabetologia. 2024. PMID: 38409439
-
NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer.Nat Commun. 2018 Aug 6;9(1):3112. doi: 10.1038/s41467-018-05582-x. Nat Commun. 2018. PMID: 30082712 Free PMC article.
References
-
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. - PubMed
-
- Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. - PubMed
-
- Diez J., Park Y., Zeller M., Brown D., Garza D., Ricordi C., Hutton J., Eisenbarth G.S., Pugliese A. Differential splicing of the IA-2 mRNA in pancreas and lymphoid organs as a permissive genetic mechanism for autoimmunity against the IA-2 type 1 diabetes autoantigen. Diabetes. 2001;50:895–900. - PubMed
-
- Yang F., Chen I.H., Xiong Z., Yan Y., Wang H., Yang X.F. Model of stimulation-responsive splicing and strategies in identification of immunogenic isoforms of tumor antigens and autoantigens. Clin. Immunol. 2006;121:121–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

