Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells
- PMID: 25251398
- PMCID: PMC4175465
- DOI: 10.1371/journal.pone.0107722
Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells
Abstract
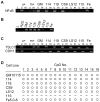
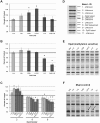
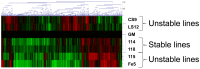
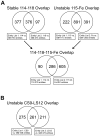
Radiation induced genomic instability is a well-studied phenomenon, the underlying mechanisms of which are poorly understood. Persistent oxidative stress, mitochondrial dysfunction, elevated cytokine levels and epigenetic changes are among the mechanisms invoked in the perpetuation of the phenotype. To determine whether epigenetic aberrations affect genomic instability we measured DNA methylation, mRNA and microRNA (miR) levels in well characterized chromosomally stable and unstable clonally expanded single cell survivors of irradiation. While no changes in DNA methylation were observed for the gene promoters evaluated, increased LINE-1 methylation was observed for two unstable clones (LS12 and CS9) and decreased Alu element methylation was observed for the other two unstable clones (115 and Fe5.0-8). These relationships also manifested for mRNA and miR expression. mRNA identified for the LS12 and CS9 clones were most similar to each other (261 mRNA), while the 115 and Fe5.0-8 clones were more similar to each other, and surprisingly also similar to the two stable clones, 114 and 118 (286 mRNA among these four clones). Pathway analysis showed enrichment for pathways involved in mitochondrial function and cellular redox, themes routinely invoked in genomic instability. The commonalities between the two subgroups of clones were also observed for miR. The number of miR for which anti-correlated mRNA were identified suggests that these miR exert functional effects in each clone. The results demonstrate significant genetic and epigenetic changes in unstable cells, but similar changes are almost as equally common in chromosomally stable cells. Possible conclusions might be that the chromosomally stable clones have some other form of instability, or that some of the observed changes represent a sort of radiation signature and that other changes are related to genomic instability. Irrespective, these findings again suggest that a spectrum of changes both drive genomic instability and permit unstable cells to persist and proliferate.
Conflict of interest statement
Figures

References
-
- Coates PJ, Lorimore SA, Wright EG (2004) Damaging and protective cell signalling in the untargeted effects of ionizing radiation. Mutation research 568: 5–20. - PubMed
-
- Little JB (2000) Radiation carcinogenesis. Carcinogenesis 21: 397–404. - PubMed
-
- Aypar U, Morgan WF, Baulch JE (2011) Radiation-induced genomic instability: are epigenetic mechanisms the missing link? International journal of radiation biology 87: 179–191. - PubMed
-
- Kovalchuk O, Baulch JE (2008) Epigenetic changes and nontargeted radiation effects–is there a link? Environ Mol Mutagen 49: 16–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

