Transcription-associated mutagenesis
- PMID: 25251854
- PMCID: PMC9464043
- DOI: 10.1146/annurev-genet-120213-092015
Transcription-associated mutagenesis
Abstract
Transcription requires unwinding complementary DNA strands, generating torsional stress, and sensitizing the exposed single strands to chemical reactions and endogenous damaging agents. In addition, transcription can occur concomitantly with the other major DNA metabolic processes (replication, repair, and recombination), creating opportunities for either cooperation or conflict. Genetic modifications associated with transcription are a global issue in the small genomes of microorganisms in which noncoding sequences are rare. Transcription likewise becomes significant when one considers that most of the human genome is transcriptionally active. In this review, we focus specifically on the mutagenic consequences of transcription. Mechanisms of transcription-associated mutagenesis in microorganisms are discussed, as is the role of transcription in somatic instability of the vertebrate immune system.
Keywords: DNA damage; class-switch recombination; cytosine deamination; somatic hypermutation; topoisomerase.
Figures

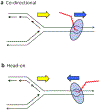

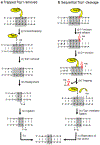
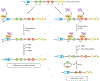
References
-
- Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46:115–24 - PubMed
-
- Arakawa H, Hauschild J, Buerstedde JM. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301–6 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

