Single-cell analyses of regulatory network perturbations using enhancer-targeting TALEs suggest novel roles for PU.1 during haematopoietic specification
- PMID: 25252941
- PMCID: PMC4197694
- DOI: 10.1242/dev.115709
Single-cell analyses of regulatory network perturbations using enhancer-targeting TALEs suggest novel roles for PU.1 during haematopoietic specification
Abstract
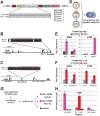
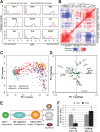
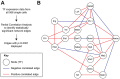
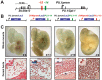
Transcription factors (TFs) act within wider regulatory networks to control cell identity and fate. Numerous TFs, including Scl (Tal1) and PU.1 (Spi1), are known regulators of developmental and adult haematopoiesis, but how they act within wider TF networks is still poorly understood. Transcription activator-like effectors (TALEs) are a novel class of genetic tool based on the modular DNA-binding domains of Xanthomonas TAL proteins, which enable DNA sequence-specific targeting and the manipulation of endogenous gene expression. Here, we report TALEs engineered to target the PU.1-14kb and Scl+40kb transcriptional enhancers as efficient new tools to perturb the expression of these key haematopoietic TFs. We confirmed the efficiency of these TALEs at the single-cell level using high-throughput RT-qPCR, which also allowed us to assess the consequences of both PU.1 activation and repression on wider TF networks during developmental haematopoiesis. Combined with comprehensive cellular assays, these experiments uncovered novel roles for PU.1 during early haematopoietic specification. Finally, transgenic mouse studies confirmed that the PU.1-14kb element is active at sites of definitive haematopoiesis in vivo and PU.1 is detectable in haemogenic endothelium and early committing blood cells. We therefore establish TALEs as powerful new tools to study the functionality of transcriptional networks that control developmental processes such as early haematopoiesis.
Keywords: Haematopoiesis; PU.1; Regulatory networks; Transcription activator-like effectors.
© 2014. Published by The Company of Biologists Ltd.
Figures



References
-
- Bailey, N. T. J. (1995). Statistical Methods in Biology. Cambridge, UK: Cambridge University Press.
-
- Beerli, R. R., Segal, D. J., Dreier, B. and Barbas, C. F., III (1998). Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 95, 14628-14633 10.1073/pnas.95.25.14628 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12009/2/MRC_/Medical Research Council/United Kingdom
- 12765/CRUK_/Cancer Research UK/United Kingdom
- RG/09/012/28096/BHF_/British Heart Foundation/United Kingdom
- BB/I00050X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- 100140/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- RP-PG-0310-1002/DH_/Department of Health/United Kingdom
- C1163/A12765/WT_/Wellcome Trust/United Kingdom
- G1000801H/MRC_/Medical Research Council/United Kingdom
- G0900951/MRC_/Medical Research Council/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

