Rb suppresses human cone-precursor-derived retinoblastoma tumours
- PMID: 25252974
- PMCID: PMC4232224
- DOI: 10.1038/nature13813
Rb suppresses human cone-precursor-derived retinoblastoma tumours
Abstract
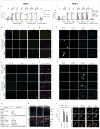
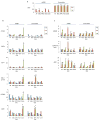
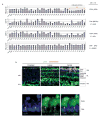
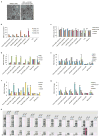
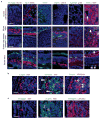
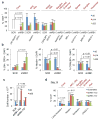
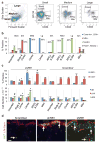
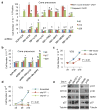
Retinoblastoma is a childhood retinal tumour that initiates in response to biallelic RB1 inactivation and loss of functional retinoblastoma (Rb) protein. Although Rb has diverse tumour-suppressor functions and is inactivated in many cancers, germline RB1 mutations predispose to retinoblastoma far more strongly than to other malignancies. This tropism suggests that retinal cell-type-specific circuitry sensitizes to Rb loss, yet the nature of the circuitry and the cell type in which it operates have been unclear. Here we show that post-mitotic human cone precursors are uniquely sensitive to Rb depletion. Rb knockdown induced cone precursor proliferation in prospectively isolated populations and in intact retina. Proliferation followed the induction of E2F-regulated genes, and depended on factors having strong expression in maturing cone precursors and crucial roles in retinoblastoma cell proliferation, including MYCN and MDM2. Proliferation of Rb-depleted cones and retinoblastoma cells also depended on the Rb-related protein p107, SKP2, and a p27 downregulation associated with cone precursor maturation. Moreover, Rb-depleted cone precursors formed tumours in orthotopic xenografts with histological features and protein expression typical of human retinoblastoma. These findings provide a compelling molecular rationale for a cone precursor origin of retinoblastoma. More generally, they demonstrate that cell-type-specific circuitry can collaborate with an initiating oncogenic mutation to enable tumorigenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






Comment in
-
Cancer: The origin of human retinoblastoma.Nature. 2014 Oct 16;514(7522):312-3. doi: 10.1038/nature13748. Epub 2014 Sep 24. Nature. 2014. PMID: 25252972 Free PMC article.
-
Tumorigenesis: establishing the origin of retinoblastoma.Nat Rev Cancer. 2014 Nov;14(11):706-7. doi: 10.1038/nrc3849. Epub 2014 Oct 16. Nat Rev Cancer. 2014. PMID: 25319865 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

