Turnover and accumulation of genetic diversity across large time-scale cycles of isolation and connection of populations
- PMID: 25253456
- PMCID: PMC4211446
- DOI: 10.1098/rspb.2014.1369
Turnover and accumulation of genetic diversity across large time-scale cycles of isolation and connection of populations
Abstract
Major climatic and geological events but also population history (secondary contacts) have generated cycles of population isolation and connection of long and short periods. Recent empirical and theoretical studies suggest that fast evolutionary processes might be triggered by such events, as commonly illustrated in ecology by the adaptive radiation of cichlid fishes (isolation and reconnection of lakes and watersheds) and in epidemiology by the fast adaptation of the influenza virus (isolation and reconnection in hosts). We test whether cyclic population isolation and connection provide the raw material (standing genetic variation) for species evolution and diversification. Our analytical results demonstrate that population isolation and connection can provide, to populations, a high excess of genetic diversity compared with what is expected at equilibrium. This excess is either cyclic (high allele turnover) or cumulates with time depending on the duration of the isolation and the connection periods and the mutation rate. We show that diversification rates of animal clades are associated with specific periods of climatic cycles in the Quaternary. We finally discuss the importance of our results for macroevolutionary patterns and for the inference of population history from genomic data.
Keywords: diversification; migration; population subdivision; secondary contact.
© 2014 The Author(s) Published by the Royal Society. All rights reserved.
Figures
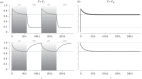
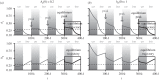

 , 2N = 1000 for each population, 200 000 generations. (d) P and net diversification rate for species representative of the main animal orders: mammals (Muridae, Ursidae, Bovidae, Cervidae, Hystricidae, Cercopithecidae, Cebidae, Cricetidae [42]), birds (passerines, [43], Darwin's finches, [44]), reptiles (collubrid snakes, [42], annoline lizards, [45]), bony fishes (Ictalurus, Cyprinidae, [42], cichlids, [46]), insects (Tipulidae, Formicinae [42]), bivalves (Petricolidae, Mesodematidae, Semelidae [42]), barnacles [42] and echinoids (Mellitidae, Clypeasteridae, Laganidae [42]). P corresponds to the period of climatic cycles in generations (100 000 years cycles divided by the generation time of each species). The shaded area represents the range of diversification rates, computed in sliding windows of 0.2 log10(years) (see alternative window sizes in the electronic supplementary material, figure S1). The dashed and dotted lines represent the values of PI for a gene of 1 and 10 kb (haplotype blocks in Eukaryotes range from 10 kb to hundreds of kb [47]), respectively, and a mutation rate of 10−8 bp−1 (mutation rates in animals range from 10−9 bp−1 to 5.10−8 bp−1 [48]).
, 2N = 1000 for each population, 200 000 generations. (d) P and net diversification rate for species representative of the main animal orders: mammals (Muridae, Ursidae, Bovidae, Cervidae, Hystricidae, Cercopithecidae, Cebidae, Cricetidae [42]), birds (passerines, [43], Darwin's finches, [44]), reptiles (collubrid snakes, [42], annoline lizards, [45]), bony fishes (Ictalurus, Cyprinidae, [42], cichlids, [46]), insects (Tipulidae, Formicinae [42]), bivalves (Petricolidae, Mesodematidae, Semelidae [42]), barnacles [42] and echinoids (Mellitidae, Clypeasteridae, Laganidae [42]). P corresponds to the period of climatic cycles in generations (100 000 years cycles divided by the generation time of each species). The shaded area represents the range of diversification rates, computed in sliding windows of 0.2 log10(years) (see alternative window sizes in the electronic supplementary material, figure S1). The dashed and dotted lines represent the values of PI for a gene of 1 and 10 kb (haplotype blocks in Eukaryotes range from 10 kb to hundreds of kb [47]), respectively, and a mutation rate of 10−8 bp−1 (mutation rates in animals range from 10−9 bp−1 to 5.10−8 bp−1 [48]).References
-
- Szabo B, Haynes C, Jr, Maxwell TA. 1995. Ages of Quaternary pluvial episodes determined by uranium-series and radiocarbon dating of lacustrine deposits of eastern Sahara. Palaeogeogr. Palaeoclimatol. Palaeoecol. 113, 227–242. ( 10.1016/0031-0182(95)00052-N) - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
